8HVF
 
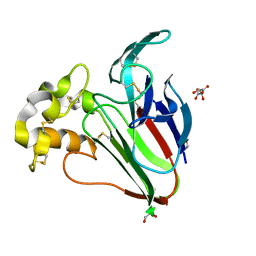 | |
8HVE
 
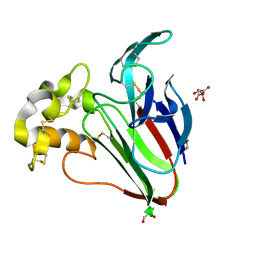 | |
8GMW
 
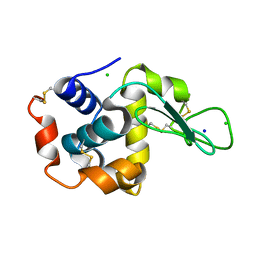 | | Crystal structure of lysozyme | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Nam, K.H. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Crystal structure of lysozyme
To Be Published
|
|
8GMV
 
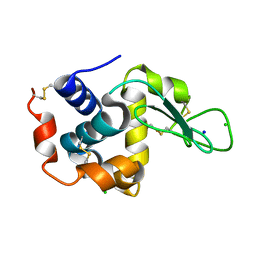 | | Crystal structure of lysozyme | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Nam, K.H. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of lysozyme
To Be Published
|
|
7WBE
 
 | |
7WBF
 
 | | Crystal structure of lysozyme | | 分子名称: | CHLORIDE ION, Lysozyme C | | 著者 | Nam, K.H. | | 登録日 | 2021-12-16 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Processing of Multicrystal Diffraction Patterns in Macromolecular Crystallography Using Serial Crystallography Programs.
Crystals, 12, 2022
|
|
7WBD
 
 | |
7XF8
 
 | |
7XF6
 
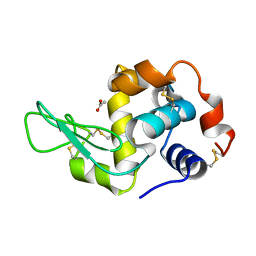 | | Crystal Structure of Human Lysozyme | | 分子名称: | ACETATE ION, Lysozyme C | | 著者 | Nam, K.H. | | 登録日 | 2022-04-01 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal Structure of Human Lysozyme Complexed with N-Acetyl-alpha-d-glucosamine.
Appl Sci (Basel), 12, 2022
|
|
7XF7
 
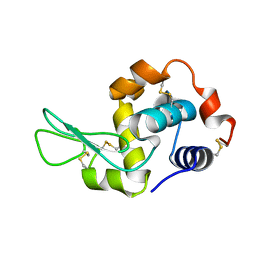 | |
8WFT
 
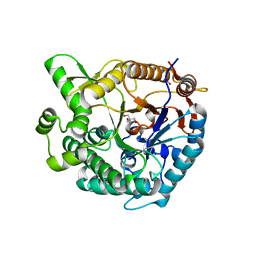 | |
8WGK
 
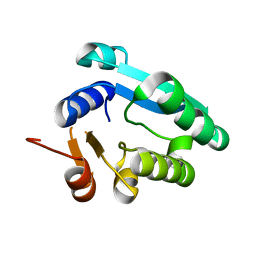 | |
8WFW
 
 | |
8WFU
 
 | |
8WDI
 
 | |
8WXO
 
 | |
8WGL
 
 | |
8X1D
 
 | |
8WFV
 
 | |
8WXM
 
 | |
8WXN
 
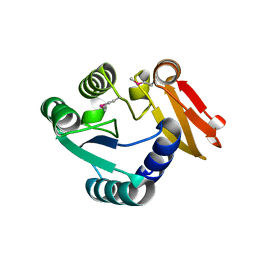 | |
8WXP
 
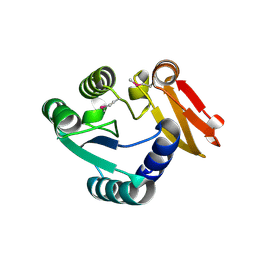 | |
8XPE
 
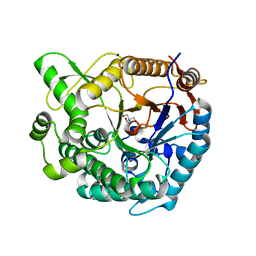 | | Crystal structure of Tris-bound TsaBgl (DATA III) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | 著者 | Nam, K.H. | | 登録日 | 2024-01-03 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural analysis of Tris binding in beta-glucosidases.
Biochem.Biophys.Res.Commun., 700, 2024
|
|
8XPC
 
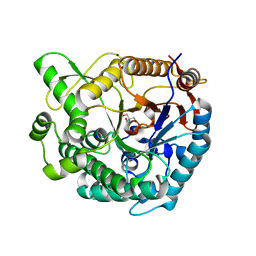 | | Crystal structure of Tris-bound TsaBgl (DATA I) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | 著者 | Nam, K.H. | | 登録日 | 2024-01-03 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural analysis of Tris binding in beta-glucosidases.
Biochem.Biophys.Res.Commun., 700, 2024
|
|
8XPD
 
 | | Crystal structure of Tris-bound TsaBgl (DATA II) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, beta-glucosidase | | 著者 | Nam, K.H. | | 登録日 | 2024-01-03 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural analysis of Tris binding in beta-glucosidases.
Biochem.Biophys.Res.Commun., 700, 2024
|
|
