7RC3
 
 | | Aeronamide N-methyltransferase, AerE (Y137F) | | Descriptor: | ASPARTIC ACID, CALCIUM ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RC2
 
 | | Aeronamide N-methyltransferase, AerE | | Descriptor: | CALCIUM ION, Methyltransferase family protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RC5
 
 | | Aeronamide N-methyltransferase, AerE (N231A) | | Descriptor: | CALCIUM ION, Methyltransferase family protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RC4
 
 | | Aeronamide N-methyltransferase, AerE (D141A) | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, Methyltransferase family protein, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RC6
 
 | | Aeronamide N-methyltransferase, AerE, bound to modified peptide substrate, AerA-DL,34 | | Descriptor: | Aeronamide A peptide, Methyltransferase family protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Cogan, D.P, Reyes, R, Nair, S.K. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure and mechanism for iterative amide N -methylation in the biosynthesis of channel-forming peptide cytotoxins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4NEI
 
 | | Alg17c PL17 Family Alginate Lyase | | Descriptor: | ZINC ION, alginate lyase | | Authors: | Park, D.S, Nair, S.K. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a PL17 Family Alginate Lyase Demonstrates Functional Similarities among Exotype Depolymerases.
J.Biol.Chem., 289, 2014
|
|
4ETX
 
 | |
4EUV
 
 | | Crystal Structure of PelD 158-CT from Pseudomonas aeruginosa PAO1, in complex with c-di-GMP, form 1 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), PelD | | Authors: | Li, Z, Chen, J, Nair, S.K. | | Deposit date: | 2012-04-25 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the PelD Cyclic Diguanylate Effector Involved in Pellicle Formation in Pseudomonas aeruginosa PAO1.
J.Biol.Chem., 287, 2012
|
|
4ETW
 
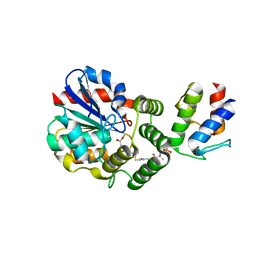 | | Structure of the Enzyme-ACP Substrate Gatekeeper Complex Required for Biotin Synthesis | | Descriptor: | Acyl carrier protein, Pimelyl-[acyl-carrier protein] methyl ester esterase, methyl 7-{[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-7-oxoheptanoate | | Authors: | Agarwal, V, Nair, S.K. | | Deposit date: | 2012-04-24 | | Release date: | 2012-10-17 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the enzyme-acyl carrier protein (ACP) substrate gatekeeper complex required for biotin synthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4QEC
 
 | | ElxO with NADP Bound | | Descriptor: | ElxO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2014-05-15 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Specificity of the Lanthipeptide Peptidase ElxP and the Oxidoreductase ElxO.
Acs Chem.Biol., 9, 2014
|
|
4EU0
 
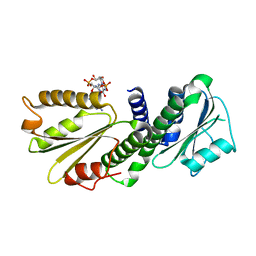 | | Crystal Structure of PelD 158-CT from Pseudomonas aeruginosa PAO1 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), PelD | | Authors: | Li, Z, Chen, J, Nair, S.K. | | Deposit date: | 2012-04-24 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the PelD Cyclic Diguanylate Effector Involved in Pellicle Formation in Pseudomonas aeruginosa PAO1.
J.Biol.Chem., 287, 2012
|
|
4Q85
 
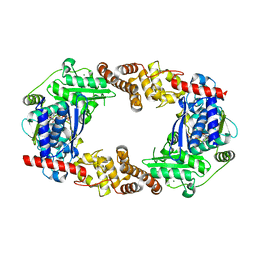 | | YcaO with Non-hydrolyzable ATP (AMPCPP) Bound | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, Ribosomal protein S12 methylthiotransferase accessory factor YcaO | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2014-04-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis.
Nat.Chem.Biol., 10, 2014
|
|
4ETZ
 
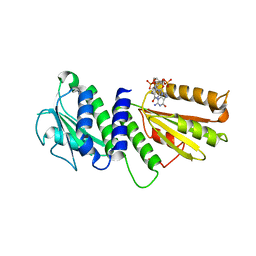 | | Crystal Structure of PelD 158-CT from Pseudomonas aeruginosa PAO1 | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), PelD | | Authors: | Li, Z, Chen, J, Nair, S.K. | | Deposit date: | 2012-04-24 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of the PelD Cyclic Diguanylate Effector Involved in Pellicle Formation in Pseudomonas aeruginosa PAO1.
J.Biol.Chem., 287, 2012
|
|
4Q84
 
 | | Apo YcaO | | Descriptor: | MERCURY (II) ION, Ribosomal protein S12 methylthiotransferase accessory factor YcaO | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2014-04-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis.
Nat.Chem.Biol., 10, 2014
|
|
4Q86
 
 | | YcaO with AMP Bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Ribosomal protein S12 methylthiotransferase accessory factor YcaO | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2014-04-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis.
Nat.Chem.Biol., 10, 2014
|
|
4QED
 
 | | ElxO Y152F with NADPH Bound | | Descriptor: | ElxO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2014-05-15 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Specificity of the Lanthipeptide Peptidase ElxP and the Oxidoreductase ElxO.
Acs Chem.Biol., 9, 2014
|
|
4R9F
 
 | | CpMnBP1 with Mannobiose Bound | | Descriptor: | MBP1, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Chekan, J.R, Agarwal, V, Nair, S.K. | | Deposit date: | 2014-09-04 | | Release date: | 2014-10-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Basis for Mannan Utilization by Caldanaerobius polysaccharolyticus Strain ATCC BAA-17.
J.Biol.Chem., 289, 2014
|
|
4R9G
 
 | | CpMnBP1 with Mannotriose Bound | | Descriptor: | MBP1, ZINC ION, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Chekan, J.R, Agarwal, V, Nair, S.K. | | Deposit date: | 2014-09-04 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Biochemical Basis for Mannan Utilization by Caldanaerobius polysaccharolyticus Strain ATCC BAA-17.
J.Biol.Chem., 289, 2014
|
|
4QPW
 
 | | BiXyn10A CBM1 with Xylohexaose Bound | | Descriptor: | beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, glycosyl hydrolase family 10 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2014-06-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4G68
 
 | |
4GLP
 
 | |
4GRC
 
 | | Crystal structure of DyP-type peroxidase (SCO2276) from Streptomyces coelicolor | | Descriptor: | CALCIUM ION, HYDROGEN PEROXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lukk, T, Hetta, A.M.A, Jones, A, Solbiati, J, Majumdar, S, Cronan, J.E, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DyP-type peroxidases from Stretptomyces and Thermobifida can modify organosolv lignin.
To be Published
|
|
4GT2
 
 | | Crystal structure of DyP-type peroxidase (SCO3963) from Streptomyces coelicolor | | Descriptor: | ACETATE ION, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Lukk, T, Hetta, A.M.A, Jones, A, Solbiati, J, Majumdar, S, Cronan, J.E, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2012-08-28 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DyP-type peroxidases from Stretptomyces and Thermobifida can modify organosolv lignin.
To be Published
|
|
4GU7
 
 | | Crystal structure of DyP-type peroxidase (SCO7193) from Streptomyces coelicolor | | Descriptor: | NICKEL (II) ION, PROTOPORPHYRIN IX CONTAINING FE, Putative uncharacterized protein SCO7193 | | Authors: | Lukk, T, Hetta, A.M.A, Jones, A, Solbiati, J, Majumdar, S, Cronan, J.E, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2012-08-29 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | DyP-type peroxidases from Stretptomyces and Thermobifida can modify organosolv lignin.
To be Published
|
|
4GYP
 
 | | Crystal structure of the heterotetrameric complex of GlucD and GlucDRP from E. coli K-12 MG1655 (EFI TARGET EFI-506058) | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Lukk, T, Ghasempur, S, Imker, H.J, Gerlt, J.A, Nair, S.K, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glucarate dehydratase and its related protein from Escherichia coli form a heterotetrameric complex.
to be published
|
|
