3UQ5
 
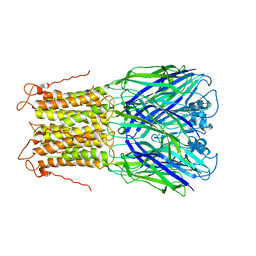 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) mutant L240A F247L (L9A F16L) in the presence of 10 mM cysteamine | | 分子名称: | Gamma-aminobutyric-acid receptor subunit beta-1, SODIUM ION | | 著者 | Gonzalez-Gutierrez, G, Lukk, T, Agarwal, V, Papke, D, Nair, S.K, Grosman, C. | | 登録日 | 2011-11-19 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UQ7
 
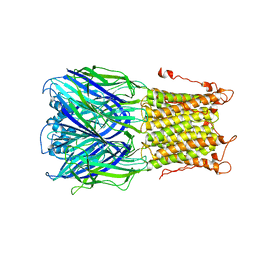 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) mutant L240S F247L (L9S F16L) in presence of 10 mM cysteamine | | 分子名称: | Gamma-aminobutyric-acid receptor subunit beta-1 | | 著者 | Gonzalez-Gutierrez, G, Lukk, T, Agarwal, V, Papke, D, Nair, S.K, Grosman, C. | | 登録日 | 2011-11-19 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2NUK
 
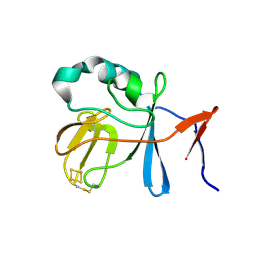 | | Soluble Domain of the Rieske Iron-Sulfur Protein from Rhodobacter sphaeroides | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | 著者 | Kolling, D, Brunzelle, J, Lhee, S, Crofts, A.R, Nair, S.K. | | 登録日 | 2006-11-09 | | 公開日 | 2007-04-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
2NVF
 
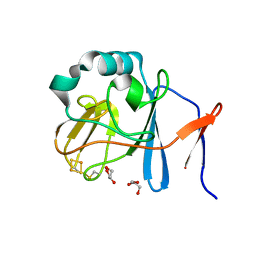 | | Soluble domain of Rieske Iron-Sulfur protein. | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | 著者 | Kolling, D, Brunzelle, J, Lhee, S, Crofts, A.R, Nair, S.K. | | 登録日 | 2006-11-12 | | 公開日 | 2007-02-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
2NWF
 
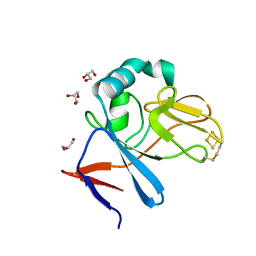 | | Soluble domain of Rieske Iron Sulfur Protein | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | 著者 | Kolling, D, Brunzelle, J.S, Lhee, S, Crofts, A.R, Nair, S.K. | | 登録日 | 2006-11-14 | | 公開日 | 2007-04-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
2NUM
 
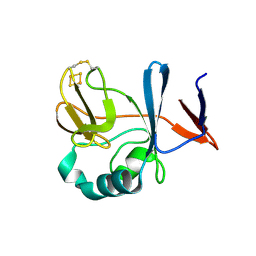 | | Soluble domain of Rieske Iron-Sulfur Protein | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | 著者 | Kolling, D, Brunzelle, J, Lhee, S, Crofts, A.R, Nair, S.K. | | 登録日 | 2006-11-09 | | 公開日 | 2007-02-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
2OKV
 
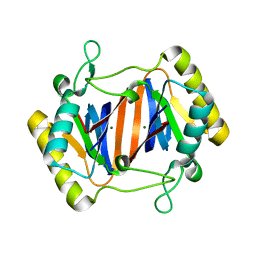 | | c-Myc DNA Unwinding Element Binding Protein | | 分子名称: | MAGNESIUM ION, Probable D-tyrosyl-tRNA(Tyr) deacylase 1 | | 著者 | Bae, B, Nair, S.K. | | 登録日 | 2007-01-17 | | 公開日 | 2007-01-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and Function of the c-myc DNA-unwinding Element-binding Protein DUE-B.
J.Biol.Chem., 282, 2007
|
|
2NVE
 
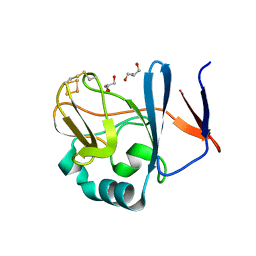 | | Soluble domain of Rieske Iron Sulfur Protein | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | 著者 | Kolling, D.K, Brunzelle, J.S, Lhee, S, Crofts, A.R, Nair, S.K. | | 登録日 | 2006-11-12 | | 公開日 | 2007-02-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
2NVG
 
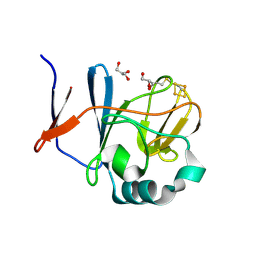 | | Soluble domain of Rieske Iron Sulfur protein. | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, Ubiquinol-cytochrome c reductase iron-sulfur subunit | | 著者 | Kolling, D, Brunzelle, J, Lhee, S, Crofts, A.R, Nair, S.K. | | 登録日 | 2006-11-12 | | 公開日 | 2007-02-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Atomic resolution structures of rieske iron-sulfur protein: role of hydrogen bonds in tuning the redox potential of iron-sulfur clusters.
Structure, 15, 2007
|
|
3JPU
 
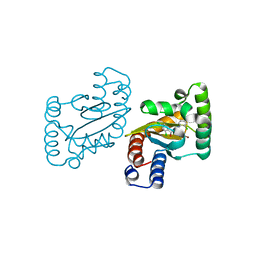 | | LasR-TP4 complex | | 分子名称: | 4-bromo-2-({[(2-chlorophenyl)carbonyl]amino}methyl)-6-methylphenyl 2,4-dichlorobenzoate, Transcriptional activator protein lasR | | 著者 | Zou, Y, Nair, S.K. | | 登録日 | 2009-09-04 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor.
Chem.Biol., 16, 2009
|
|
3M6I
 
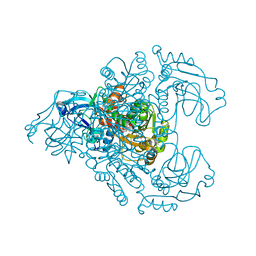 | | L-arabinitol 4-dehydrogenase | | 分子名称: | L-arabinitol 4-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | 著者 | Bae, B, Nair, S.K. | | 登録日 | 2010-03-15 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure and engineering of L-arabinitol 4-dehydrogenase from Neurospora crassa
J.Mol.Biol., 402, 2010
|
|
3OEA
 
 | |
3OEB
 
 | |
3OO3
 
 | |
3OU2
 
 | | DhpI-SAH complex structure | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase | | 著者 | Bae, B, Nair, S.K. | | 登録日 | 2010-09-14 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OU7
 
 | | DhpI-SAM-HEP complex | | 分子名称: | (2-hydroxyethyl)phosphonic acid, S-ADENOSYLMETHIONINE, SAM-dependent methyltransferase, ... | | 著者 | Bae, B, Nair, S.K. | | 登録日 | 2010-09-14 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OU6
 
 | | DhpI-SAM complex | | 分子名称: | S-ADENOSYLMETHIONINE, SAM-dependent methyltransferase, SULFATE ION | | 著者 | Bae, B, Nair, S.K. | | 登録日 | 2010-09-14 | | 公開日 | 2010-10-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3EUO
 
 | |
3EUQ
 
 | |
3EUT
 
 | |
3F8T
 
 | |
3GBF
 
 | |
3IX4
 
 | | LasR-TP1 complex | | 分子名称: | 2,4-dibromo-6-({[(2-nitrophenyl)carbonyl]amino}methyl)phenyl 2-chlorobenzoate, Transcriptional activator protein lasR | | 著者 | Zou, Y, Nair, S.K. | | 登録日 | 2009-09-03 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor.
Chem.Biol., 16, 2009
|
|
3IX8
 
 | | LasR-TP3 complex | | 分子名称: | 2,4-dibromo-6-({[(2-chlorophenyl)carbonyl]amino}methyl)phenyl 2-methylbenzoate, Transcriptional activator protein lasR | | 著者 | Zou, Y, Nair, S.K. | | 登録日 | 2009-09-03 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor.
Chem.Biol., 16, 2009
|
|
3IX3
 
 | | LasR-OC12 HSL complex | | 分子名称: | N-3-OXO-DODECANOYL-L-HOMOSERINE LACTONE, Transcriptional activator protein lasR | | 著者 | Zou, Y, Nair, S.K. | | 登録日 | 2009-09-03 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | LasR-OC12 HSL complex
TO BE PUBLISHED
|
|
