4HT7
 
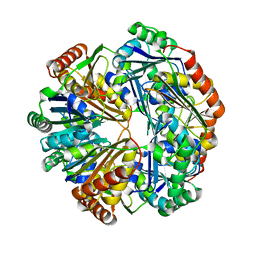 | | CO2 concentrating mechanism protein P, CcmP form 2 | | Descriptor: | CO2 concentrating mechanism protein P | | Authors: | Cai, F, Sutter, M, Kerfeld, C.A. | | Deposit date: | 2012-10-31 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | The structure of CcmP, a tandem bacterial microcompartment domain protein from the beta-carboxysome, forms a subcompartment within a microcompartment.
J.Biol.Chem., 288, 2013
|
|
4HT5
 
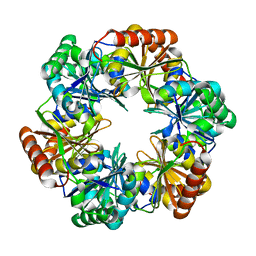 | | CO2 concentrating mechanism protein P, CcmP form 1 | | Descriptor: | CO2 concentrating mechanism protein P | | Authors: | Cai, F, Sutter, M, Kerfeld, C.A. | | Deposit date: | 2012-10-31 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The structure of CcmP, a tandem bacterial microcompartment domain protein from the beta-carboxysome, forms a subcompartment within a microcompartment.
J.Biol.Chem., 288, 2013
|
|
4JVZ
 
 | | Structure of Thermosynechococcus elongatus CcmL | | Descriptor: | Carbon dioxide concentrating mechanism protein, SULFATE ION | | Authors: | Sutter, M, Kerfeld, C.A. | | Deposit date: | 2013-03-26 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Two new high-resolution crystal structures of carboxysome pentamer proteins reveal high structural conservation of CcmL orthologs among distantly related cyanobacterial species.
Photosynth.Res., 118, 2013
|
|
4JDX
 
 | |
4JDQ
 
 | |
4JW0
 
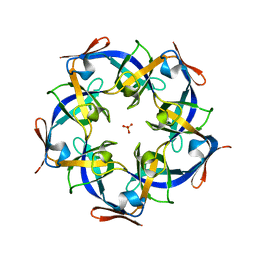 | | Structure of Gloeobacter violaceus CcmL | | Descriptor: | Carbon dioxide concentrating mechanism protein, SULFATE ION | | Authors: | Sutter, M, Kerfeld, C.A. | | Deposit date: | 2013-03-26 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two new high-resolution crystal structures of carboxysome pentamer proteins reveal high structural conservation of CcmL orthologs among distantly related cyanobacterial species.
Photosynth.Res., 118, 2013
|
|
4L7Z
 
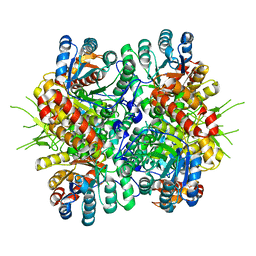 | | Crystal Structure of Chloroflexus aurantiacus malyl-CoA lyase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HpcH/HpaI aldolase | | Authors: | Zarzycki, J, Kerfeld, C.A. | | Deposit date: | 2013-06-14 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | The crystal structures of the tri-functional Chloroflexus aurantiacus and bi-functional Rhodobacter sphaeroides malyl-CoA lyases and comparison with CitE-like superfamily enzymes and malate synthases.
Bmc Struct.Biol., 13, 2013
|
|
4L9Y
 
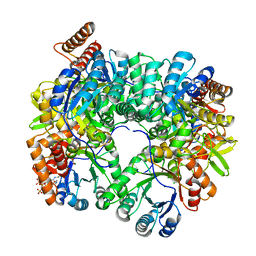 | | Crystal Structure of Rhodobacter sphaeroides malyl-CoA lyase in complex with magnesium, glyoxylate, and propionyl-CoA | | Descriptor: | CHLORIDE ION, GLYOXYLIC ACID, MAGNESIUM ION, ... | | Authors: | Zarzycki, J, Kerfeld, C.A. | | Deposit date: | 2013-06-18 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | The crystal structures of the tri-functional Chloroflexus aurantiacus and bi-functional Rhodobacter sphaeroides malyl-CoA lyases and comparison with CitE-like superfamily enzymes and malate synthases.
Bmc Struct.Biol., 13, 2013
|
|
4L9Z
 
 | | Crystal Structure of Rhodobacter sphaeroides malyl-CoA lyase in complex with magnesium, oxalate, and CoA | | Descriptor: | COENZYME A, MAGNESIUM ION, Malyl-CoA lyase, ... | | Authors: | Zarzycki, J, Kerfeld, C.A. | | Deposit date: | 2013-06-18 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | The crystal structures of the tri-functional Chloroflexus aurantiacus and bi-functional Rhodobacter sphaeroides malyl-CoA lyases and comparison with CitE-like superfamily enzymes and malate synthases.
Bmc Struct.Biol., 13, 2013
|
|
4L80
 
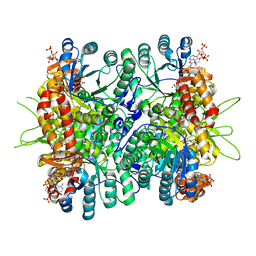 | | Crystal Structure of Chloroflexus aurantiacus malyl-CoA lyase in complex with magnesium, oxalate, and propionyl-CoA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HpcH/HpaI aldolase, MAGNESIUM ION, ... | | Authors: | Zarzycki, J, Kerfeld, C.A. | | Deposit date: | 2013-06-15 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | The crystal structures of the tri-functional Chloroflexus aurantiacus and bi-functional Rhodobacter sphaeroides malyl-CoA lyases and comparison with CitE-like superfamily enzymes and malate synthases.
Bmc Struct.Biol., 13, 2013
|
|
6MZV
 
 | |
6MZX
 
 | |
6N07
 
 | |
6N06
 
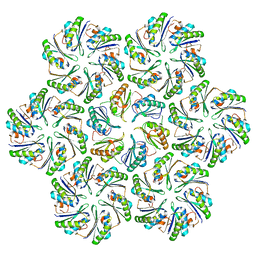 | |
6N0F
 
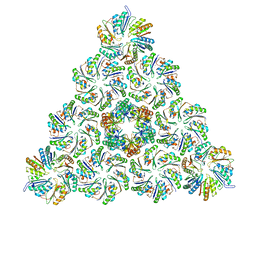 | |
6N0G
 
 | |
6MZY
 
 | |
6MZU
 
 | |
6N09
 
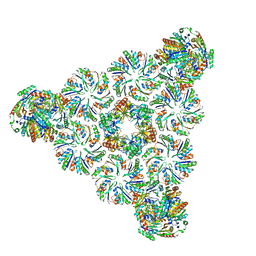 | |
6NER
 
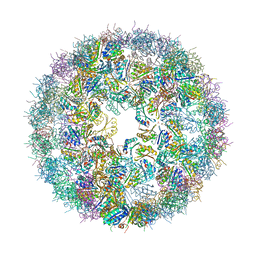 | | Synthetic Haliangium ochraceum BMC shell | | Descriptor: | BMC-H tandem fusion protein, SULFATE ION | | Authors: | Sutter, M, McGuire, S, Aussignargues, C, Kerfeld, C.A. | | Deposit date: | 2018-12-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural Characterization of a Synthetic Tandem-Domain Bacterial Microcompartment Shell Protein Capable of Forming Icosahedral Shell Assemblies.
ACS Synth Biol, 8, 2019
|
|
6NLU
 
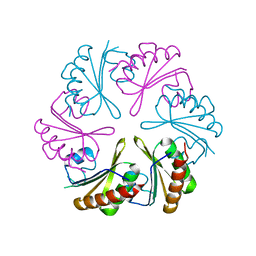 | |
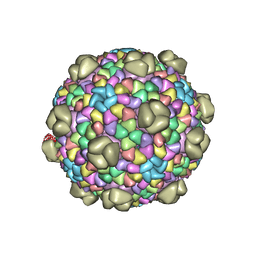
6OWG
 
 | | Structure of a synthetic beta-carboxysome shell, T=4 | | Descriptor: | Ethanolamine utilization protein EutN/carboxysome structural protein Ccml, Microcompartments protein | | Authors: | Sutter, M, Laughlin, T.G, Davies, K.M, Kerfeld, C.A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of a Syntheticbeta-Carboxysome Shell.
Plant Physiol., 181, 2019
|
|
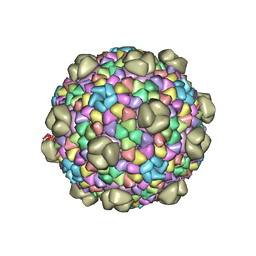
6OWF
 
 | | Structure of a synthetic beta-carboxysome shell, T=3 | | Descriptor: | Ethanolamine utilization protein EutN/carboxysome structural protein Ccml, Microcompartments protein | | Authors: | Sutter, M, Laughlin, T.G, Davies, K.M, Kerfeld, C.A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a Syntheticbeta-Carboxysome Shell.
Plant Physiol., 181, 2019
|
|
5TZ0
 
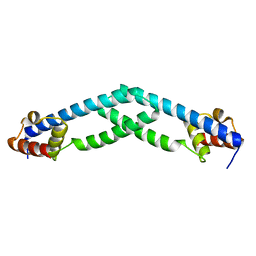 | |
5V76
 
 | | Structure of Haliangium ochraceum BMC-T HO-3341 | | Descriptor: | GLYCEROL, Microcompartments protein | | Authors: | Sutter, M, Paasch, B, Zarzycki, J, Aussignargues, C, Kerfeld, C.A. | | Deposit date: | 2017-03-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell.
Science, 356, 2017
|
|
