5FFF
 
 | |
4R6X
 
 | |
1LWI
 
 | | 3-ALPHA-HYDROXYSTEROID/DIHYDRODIOL DEHYDROGENASE FROM RATTUS NORVEGICUS | | Descriptor: | 3-ALPHA-HYDROXYSTEROID/DIHYDRODIOL DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bennett, M.J, Schlegel, B.P, Jez, J.M, Penning, T.M, Lewis, M. | | Deposit date: | 1996-02-24 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase complexed with NADP+.
Biochemistry, 35, 1996
|
|
6GSD
 
 | | Plantago Major multifunctional oxidoreductase in complex with progesterone and NADP+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROGESTERONE, Progesterone 5-beta-reductase | | Authors: | Fellows, R, Silva, C, Russo, C.M, Lee, S.G, Jez, J.M, Chisholm, J.D, Zubieta, C, Nanao, M. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A multisubstrate reductase from Plantago major: structure-function in the short chain reductase superfamily.
Sci Rep, 8, 2018
|
|
4EPM
 
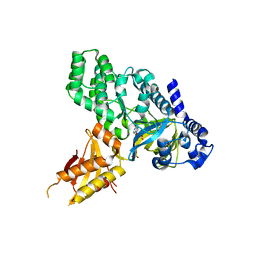 | | Crystal Structure of Arabidopsis GH3.12 (PBS3) in Complex with AMP | | Descriptor: | 4-substituted benzoates-glutamate ligase GH3.12, ADENOSINE MONOPHOSPHATE, SULFATE ION | | Authors: | Westfall, C.S, Zubieta, C, Herrmann, J, Kapp, U, Nanao, M.H, Jez, J.M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural basis for prereceptor modulation of plant hormones by GH3 proteins.
Science, 336, 2012
|
|
4EPL
 
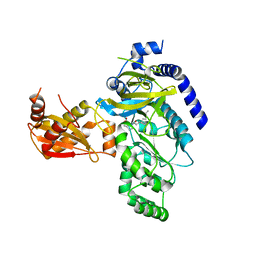 | | Crystal Structure of Arabidopsis thaliana GH3.11 (JAR1) in Complex with JA-Ile | | Descriptor: | Jasmonic acid-amido synthetase JAR1, N-({(1R,2R)-3-oxo-2-[(2Z)-pent-2-en-1-yl]cyclopentyl}acetyl)-L-isoleucine | | Authors: | Westfall, C.S, Zubieta, C, Herrmann, J, Kapp, U, Nanao, M.H, Jez, J.M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural basis for prereceptor modulation of plant hormones by GH3 proteins.
Science, 336, 2012
|
|
4S13
 
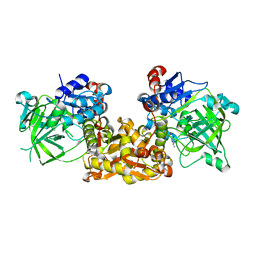 | | Ferulic Acid Decarboxylase (FDC1) | | Descriptor: | 4-ethenylphenol, Ferulic acid decarboxylase 1 | | Authors: | Lee, S.G, Bhuiya, M.W, Yu, O, Jez, J.M. | | Deposit date: | 2015-01-07 | | Release date: | 2015-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Structure and Mechanism of Ferulic Acid Decarboxylase (FDC1) from Saccharomyces cerevisiae.
Appl.Environ.Microbiol., 81, 2015
|
|
1AFS
 
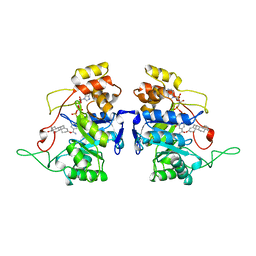 | | RECOMBINANT RAT LIVER 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE (3-ALPHA-HSD) COMPLEXED WITH NADP AND TESTOSTERONE | | Descriptor: | 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TESTOSTERONE | | Authors: | Bennett, M.J, Albert, R.H, Jez, J.M, Ma, H, Penning, T.M, Lewis, M. | | Deposit date: | 1997-03-13 | | Release date: | 1997-10-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Steroid recognition and regulation of hormone action: crystal structure of testosterone and NADP+ bound to 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase.
Structure, 5, 1997
|
|
3R8W
 
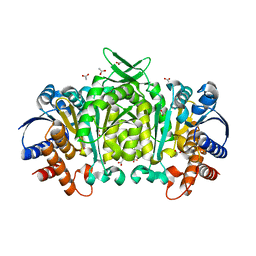 | | Structure of 3-isopropylmalate dehydrogenase isoform 2 from Arabidopsis thaliana at 2.2 angstrom resolution | | Descriptor: | 3-isopropylmalate dehydrogenase 2, chloroplastic, ACETATE ION | | Authors: | He, Y, Galant, A, Pang, Q, Strul, J.M, Balogun, S, Jez, J.M, Chen, S. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional evolution of isopropylmalate dehydrogenases in the leucine and glucosinolate pathways of Arabidopsis thaliana.
J.Biol.Chem., 286, 2011
|
|
6MH4
 
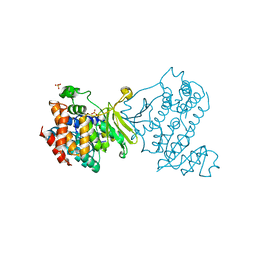 | |
6MH5
 
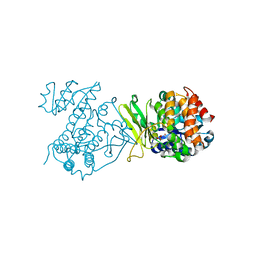 | |
7L0B
 
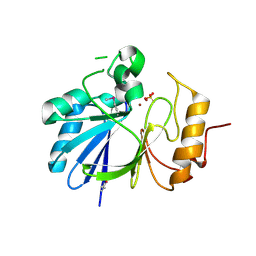 | | Crystal structure of hydroxyacyl glutathione hydrolase (GloB) from Staphylococcus aureus, apoenzyme | | Descriptor: | Hydroxyacylglutathione hydrolase, SULFATE ION, ZINC ION | | Authors: | Miller, J.J, Jez, J.M, Odom John, A.R. | | Deposit date: | 2020-12-11 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-guided microbial targeting of antistaphylococcal prodrugs.
Elife, 10, 2021
|
|
7L0A
 
 | |
6MS8
 
 | | Crystal Structure of Chalcone Isomerase from Medicago Truncatula Complexed with (2S) Naringenin | | Descriptor: | Chalcone-flavonone isomerase family protein, NARINGENIN | | Authors: | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | Deposit date: | 2018-10-16 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
3KAJ
 
 | | Apoenzyme structure of homoglutathione synthetase from Glycine max in open conformation | | Descriptor: | Homoglutathione synthetase | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
1Z7Y
 
 | | Crystal Structure of the Arabidopsis thaliana O-Acetylserine Sulfhydrylase K46A mutant | | Descriptor: | Cysteine synthase, N-[(3-HYDROXY-2-METHYL-5-{[(TRIHYDROXYPHOSPHORANYL)OXY]METHYL}PYRIDIN-4-YL)METHYLENE]METHIONINE | | Authors: | Bonner, E.R, Cahoon, R.E, Knapke, S.M, Jez, J.M. | | Deposit date: | 2005-03-28 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Basis of Cysteine Biosynthesis in Plants: STRUCTURAL AND FUNCTIONAL ANALYSIS OF O-ACETYLSERINE SULFHYDRYLASE FROM ARABIDOPSIS THALIANA.
J.Biol.Chem., 280, 2005
|
|
1QLV
 
 | |
1Z7W
 
 | | Crystal Structure of O-Acetylserine Sulfhydrylase from Arabidopsis thaliana | | Descriptor: | Cysteine synthase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Bonner, E.R, Cahoon, R.E, Knapke, S.M, Jez, J.M. | | Deposit date: | 2005-03-28 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis of Cysteine Biosynthesis in Plants: STRUCTURAL AND FUNCTIONAL ANALYSIS OF O-ACETYLSERINE SULFHYDRYLASE FROM ARABIDOPSIS THALIANA.
J.Biol.Chem., 280, 2005
|
|
6WLF
 
 | |
6AVH
 
 | | GH3.15 acyl acid amido synthetase | | Descriptor: | ADENOSINE MONOPHOSPHATE, GH3.15 acyl acid amido synthetase | | Authors: | Sherp, A.M, Jez, J.M. | | Deposit date: | 2017-09-02 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Arabidopsis thalianaGH3.15 acyl acid amido synthetase has a highly specific substrate preference for the auxin precursor indole-3-butyric acid.
J. Biol. Chem., 293, 2018
|
|
6X9L
 
 | |
6CJO
 
 | | Crystal Structure of Chalcone Isomerase from Medicago Sativa with the G95S mutation. | | Descriptor: | Chalcone--flavonone isomerase 1, SULFATE ION | | Authors: | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
7MKU
 
 | |
2ISQ
 
 | | Crystal Structure of O-Acetylserine Sulfhydrylase from Arabidopsis Thaliana in Complex with C-Terminal Peptide from Arabidopsis Serine Acetyltransferase | | Descriptor: | Cysteine synthase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION, ... | | Authors: | Francois, J.A, Kumaran, S, Jez, J.M. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for interaction of o-acetylserine sulfhydrylase and serine acetyltransferase in the Arabidopsis cysteine synthase complex.
Plant Cell, 18, 2006
|
|
5CB8
 
 | |
