2CI1
 
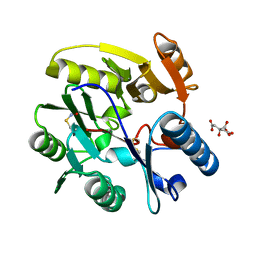 | | Crystal Structure of dimethylarginine dimethylaminohydrolase I in complex with S-nitroso-Lhomocysteine | | Descriptor: | CITRIC ACID, NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1 | | Authors: | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | Deposit date: | 2006-03-17 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inhibitors.
Structure, 14, 2006
|
|
2CI3
 
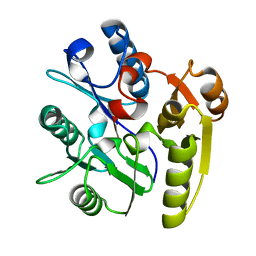 | | Crystal Structure of Dimethylarginine dimethylaminohydrolase crystal form I | | Descriptor: | NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1 | | Authors: | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | Deposit date: | 2006-03-17 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inhibitors.
Structure, 14, 2006
|
|
2CI5
 
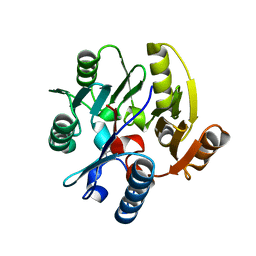 | | Crystal structure of Dimethylarginine Dimethylaminohydrolase I in complex with L-homocysteine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, CITRIC ACID, NG, ... | | Authors: | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | Deposit date: | 2006-03-17 | | Release date: | 2006-05-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inbitors
Structure, 14, 2006
|
|
2DKO
 
 | | Extended substrate recognition in caspase-3 revealed by high resolution X-ray structure analysis | | Descriptor: | Caspase-3, PHQ-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE | | Authors: | Mittl, P.R.E, Ganesan, R, Jelakovic, S, Grutter, M.G. | | Deposit date: | 2006-04-12 | | Release date: | 2006-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Extended Substrate Recognition in Caspase-3 Revealed by High Resolution X-ray Structure Analysis
J.Mol.Biol., 359, 2006
|
|
1TZE
 
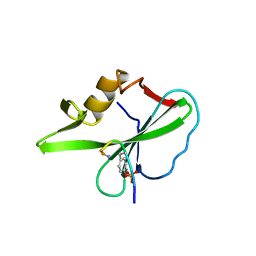 | |
1JPF
 
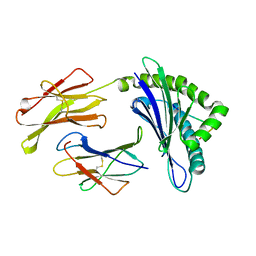 | | Crystal Structure Of The LCMV Peptidic Epitope Gp276 In Complex With The Murine Class I Mhc Molecule H-2Db | | Descriptor: | BETA-2-MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Ciatto, C, Tissot, A.C, Tschopp, M, Capitani, G, Pecorari, F, Pluckthun, A, Grutter, M.G. | | Deposit date: | 2001-08-02 | | Release date: | 2001-10-24 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Zooming in on the hydrophobic ridge of H-2D(b): implications for the conformational variability of bound peptides.
J.Mol.Biol., 312, 2001
|
|
1JPG
 
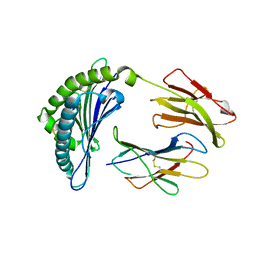 | | Crystal Structure Of The LCMV Peptidic Epitope Np396 In Complex With The Murine Class I Mhc Molecule H-2Db | | Descriptor: | BETA-2-MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Ciatto, C, Tissot, A.C, Tschopp, M, Capitani, G, Pecorari, F, Pluckthun, A, Grutter, M.G. | | Deposit date: | 2001-08-02 | | Release date: | 2001-10-24 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zooming in on the hydrophobic ridge of H-2D(b): implications for the conformational variability of bound peptides.
J.Mol.Biol., 312, 2001
|
|
1KSO
 
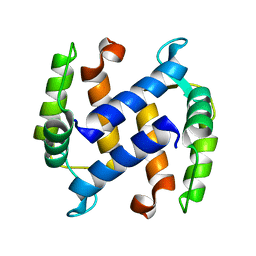 | | CRYSTAL STRUCTURE OF APO S100A3 | | Descriptor: | S100 CALCIUM-BINDING PROTEIN A3 | | Authors: | Mittl, P.R, Fritz, G, Sargent, D.F, Richmond, T.J, Heizmann, C.W, Grutter, M.G. | | Deposit date: | 2002-01-14 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal-free MIRAS phasing: structure of apo-S100A3.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KKC
 
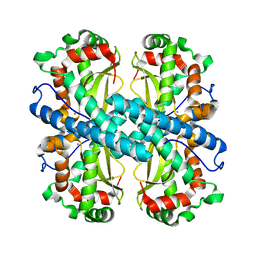 | | Crystal structure of Aspergillus fumigatus MnSOD | | Descriptor: | MANGANESE (II) ION, Manganese Superoxide Dismutase | | Authors: | Fluckiger, S, Mittl, P.R.E, Scapozza, L, Fijten, H, Folkers, G, Grutter, M.G, Blaser, K, Crameri, R. | | Deposit date: | 2001-12-07 | | Release date: | 2001-12-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparison of the crystal structures of the human manganese superoxide dismutase and the homologous Aspergillus fumigatus allergen at 2-A resolution.
J.Immunol., 168, 2002
|
|
1M4N
 
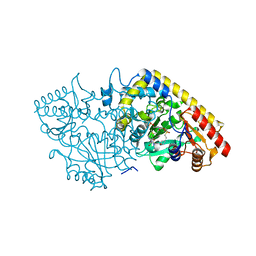 | | CRYSTAL STRUCTURE OF APPLE ACC SYNTHASE IN COMPLEX WITH [2-(AMINO-OXY)ETHYL](5'-DEOXYADENOSIN-5'-YL)(METHYL)SULFONIUM | | Descriptor: | (2-AMINOOXY-ETHYL)-[5-(6-AMINO-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDRO-FURAN-2-YLMETHYL]-METHYL-SULFONIUM, 1-aminocyclopropane-1-carboxylate synthase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Capitani, G, Eliot, A.C, Gut, H, Khomutov, R.M, Kirsch, J.F, Grutter, M.G. | | Deposit date: | 2002-07-03 | | Release date: | 2003-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of 1-aminocyclopropane-1-carboxylate synthase in complex with an amino-oxy analogue of the substrate: implications for substrate binding.
BIOCHEM.BIOPHYS.ACTA PROTEINS & PROTEOMICS, 1647, 2003
|
|
1NQ3
 
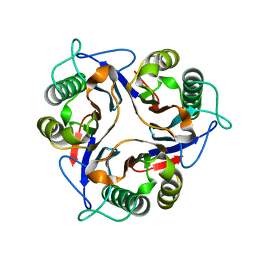 | | Crystal structure of the mammalian tumor associated antigen UK114 | | Descriptor: | 14.3 kDa perchloric acid soluble protein | | Authors: | Deriu, D, Briand, C, Mistiniene, E, Naktinis, V, Grutter, M.G. | | Deposit date: | 2003-01-21 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and oligomeric state of the mammalian tumour-associated antigen UK114.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1L10
 
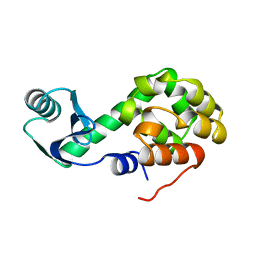 | |
1L01
 
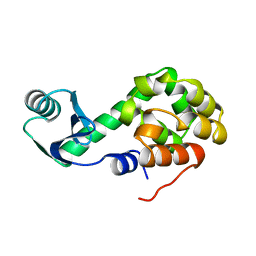 | |
4V3Q
 
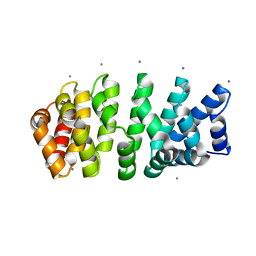 | | Designed armadillo repeat protein with 4 internal repeats, 2nd generation C-cap and 3rd generation N-cap. | | Descriptor: | CALCIUM ION, GLYCEROL, YIII_M4_AII | | Authors: | Reichen, C, Madhurantakam, C, Pluckthun, A, Mittl, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of designed armadillo-repeat proteins show propagation of inter-repeat interface effects.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1BX7
 
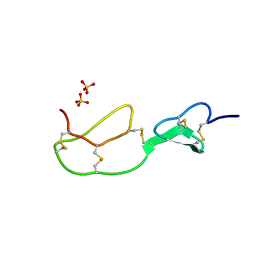 | | HIRUSTASIN FROM HIRUDO MEDICINALIS AT 1.2 ANGSTROMS | | Descriptor: | HIRUSTASIN, SULFATE ION | | Authors: | Uson, I, Sheldrick, G.M, De La Fortelle, E, Bricogne, G, Di Marco, S, Priestle, J.P, Gruetter, M.G, Mittl, P.R.E. | | Deposit date: | 1998-10-14 | | Release date: | 1999-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A crystal structure of hirustasin reveals the intrinsic flexibility of a family of highly disulphide-bridged inhibitors.
Structure Fold.Des., 7, 1999
|
|
1BX8
 
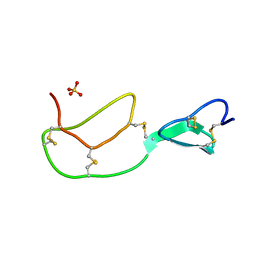 | | HIRUSTASIN FROM HIRUDO MEDICINALIS AT 1.4 ANGSTROMS | | Descriptor: | HIRUSTASIN, SULFATE ION | | Authors: | Uson, I, Sheldrick, G.M, De La Fortelle, E, Bricogne, G, Di Marco, S, Priestle, J.P, Gruetter, M.G, Mittl, P.R.E. | | Deposit date: | 1998-10-14 | | Release date: | 1999-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.2 A crystal structure of hirustasin reveals the intrinsic flexibility of a family of highly disulphide-bridged inhibitors.
Structure Fold.Des., 7, 1999
|
|
4V3R
 
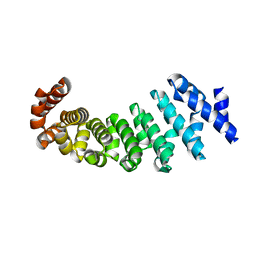 | | Designed armadillo repeat protein with 5 internal repeats, 2nd generation C-cap and 3rd generation N-cap. | | Descriptor: | MAGNESIUM ION, YIII_M5_AII | | Authors: | Reichen, C, Madhurantakam, C, Pluckthun, A, Mittl, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of Designed Armadillo-Repeat Proteins Show Propagation of Inter-Repeat Interface Effects
Acta Crystallogr.,Sect.D, 72, 2016
|
|
4V3O
 
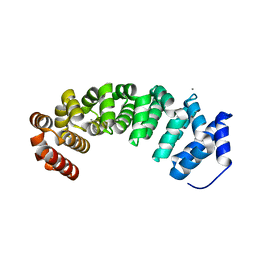 | | Designed armadillo repeat protein with 5 internal repeats, 2nd generation C-cap and 3rd generation N-cap. | | Descriptor: | ACETATE ION, CALCIUM ION, YIII_M5_AII | | Authors: | Reichen, C, Madhurantakam, C, Pluckthun, A, Mittl, P. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Designed Armadillo-Repeat Proteins Show Propagation of Inter-Repeat Interface Effects
Acta Crystallogr.,Sect.D, 72, 2016
|
|
2I1B
 
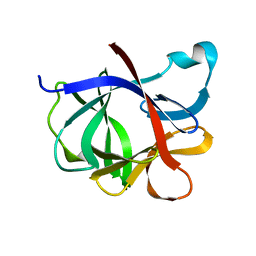 | |
1HIA
 
 | | KALLIKREIN COMPLEXED WITH HIRUSTASIN | | Descriptor: | HIRUSTASIN, KALLIKREIN | | Authors: | Mittl, P, Di Marco, S, Gruetter, M. | | Deposit date: | 1996-12-12 | | Release date: | 1997-12-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A new structural class of serine protease inhibitors revealed by the structure of the hirustasin-kallikrein complex.
Structure, 5, 1997
|
|
3QF4
 
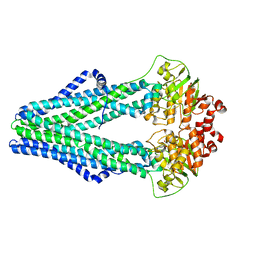 | | Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation | | Descriptor: | ABC transporter, ATP-binding protein, MAGNESIUM ION, ... | | Authors: | Hohl, M, Briand, C, Gruetter, M.G, Seeger, M.A. | | Deposit date: | 2011-01-21 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation
Nat.Struct.Mol.Biol., 19, 2012
|
|
2J8S
 
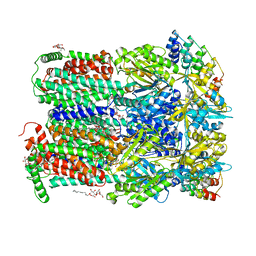 | | Drug Export Pathway of Multidrug Exporter AcrB Revealed by DARPin Inhibitors | | Descriptor: | ACRIFLAVINE RESISTANCE PROTEIN B, DARPIN, DODECYL-ALPHA-D-MALTOSIDE, ... | | Authors: | Sennhauser, G, Amstutz, P, Briand, C, Storchenegger, O, Gruetter, M.G. | | Deposit date: | 2006-10-27 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Drug Export Pathway of Multidrug Exporter Acrb Revealed by Darpin Inhibitors.
Plos Biol., 5, 2007
|
|
4Q4A
 
 | | Improved model of AMP-PNP bound TM287/288 | | Descriptor: | ABC transporter, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Hohl, M, Gruetter, M.G, Seeger, M.A. | | Deposit date: | 2014-04-14 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4Q4J
 
 | | Structure of crosslinked TM287/288_S498C_S520C mutant | | Descriptor: | ABC transporter, Uncharacterized ABC transporter ATP-binding protein TM_0288 | | Authors: | Hohl, M, Schoeppe, J, Gruetter, M.G, Seeger, M.A. | | Deposit date: | 2014-04-14 | | Release date: | 2014-07-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2K7Z
 
 | |
