1GB1
 
 | |
2GB1
 
 | |
3ZNF
 
 | |
4ZNF
 
 | |
5T82
 
 | |
4TRX
 
 | |
4FBV
 
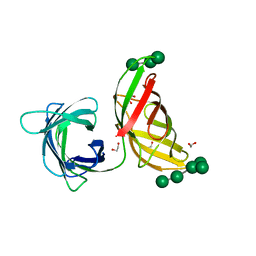 | | Crystal structure of the Myxococcus Xanthus hemagglutinin in complex with a3,a6-mannopentaose | | Descriptor: | 1,2-ETHANEDIOL, Myxobacterial hemagglutinin, alpha-D-mannopyranose, ... | | Authors: | Koharudin, L.M.I, Gronenborn, A.M. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Insights into the Anti-HIV Activity of the Oscillatoria agardhii Agglutinin Homolog Lectin Family.
J.Biol.Chem., 287, 2012
|
|
1M7T
 
 | |
9ILB
 
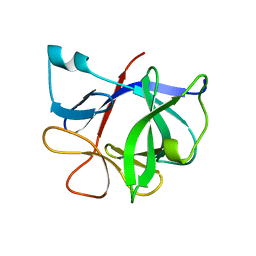 | | HUMAN INTERLEUKIN-1 BETA | | Descriptor: | PROTEIN (HUMAN INTERLEUKIN-1 BETA) | | Authors: | Yu, B, Blaber, M, Gronenborn, A.M, Clore, G.M, Caspar, D.L.D. | | Deposit date: | 1998-10-22 | | Release date: | 1999-01-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Disordered water within a hydrophobic protein cavity visualized by x-ray crystallography.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7GAT
 
 | | SOLUTION NMR STRUCTURE OF THE L22V MUTANT DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13 BP DNA CONTAINING A TGATA SITE, 34 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*AP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Leu22-->Val mutant AREA DNA binding domain complexed with a TGATAG core element defines a role for hydrophobic packing in the determination of specificity.
J.Mol.Biol., 277, 1998
|
|
6I1B
 
 | |
6WAP
 
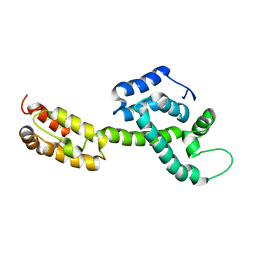 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WCY
 
 | | N160D Deamidation Mutant of Human gammaD-Crystallin | | Descriptor: | Gamma-crystallin D, SULFATE ION | | Authors: | Whitley, M.J, Rathi, N, Ambarian, M, Gronenborn, A.M. | | Deposit date: | 2020-03-31 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.204 Å) | | Cite: | Assessing the Structures and Interactions of gamma D-Crystallin Deamidation Variants.
Structure, 29, 2021
|
|
7I1B
 
 | |
2EZQ
 
 | |
2EZA
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, RESTRAINED REGULARIZED MEAN STRUCTURE | | Descriptor: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | Authors: | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | Deposit date: | 1997-05-07 | | Release date: | 1997-08-20 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
2EZC
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, 14 STRUCTURES | | Descriptor: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | Authors: | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | Deposit date: | 1997-05-07 | | Release date: | 1997-08-20 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
2EZB
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, 14 STRUCTURES | | Descriptor: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | Authors: | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | Deposit date: | 1997-05-07 | | Release date: | 1997-08-20 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
3TRX
 
 | |
6HIR
 
 | |
2EZX
 
 | |
2EZY
 
 | |
2EZZ
 
 | |
2EZD
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE SECOND DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | Authors: | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | Deposit date: | 1997-06-04 | | Release date: | 1997-10-15 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2EZM
 
 | |
