1UP2
 
 | | Structure of the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with glucose-isofagomine at 1.9 angstrom | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ACETATE ION, ... | | Authors: | Varrot, A, Leydier, S, Pell, G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-09-26 | | Release date: | 2004-11-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mycobacterium Tuberculosis Strains Possess Functional Cellulases.
J.Biol.Chem., 280, 2005
|
|
1UQY
 
 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with xylopentaose | | Descriptor: | ENDOXYLANASE, MAGNESIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose, ... | | Authors: | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
1UR1
 
 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with arabinofuranose alpha-1,3 linked to xylobiose | | Descriptor: | CHLORIDE ION, ENDOXYLANASE, MAGNESIUM ION, ... | | Authors: | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-10-24 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
1UP0
 
 | | Structure of the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with cellobiose at 1.75 angstrom | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ACETATE ION, PUTATIVE CELLULASE CEL6, ... | | Authors: | Varrot, A, Leydier, S, Pell, G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-09-26 | | Release date: | 2004-11-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mycobacterium Tuberculosis Strains Possess Functional Cellulases.
J.Biol.Chem., 280, 2005
|
|
1US2
 
 | | Xylanase10C (mutant E385A) from Cellvibrio japonicus in complex with xylopentaose | | Descriptor: | ENDO-BETA-1,4-XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Pell, G, Szabo, L, Charnock, S.J, Xie, H, Gloster, T.M, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-11-17 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Analysis of Cellvibrio Japonicus Xylanase 10C: How Variation in Substrate-Binding Cleft Influences the Catalytic Profile of Family Gh-10 Xylanases
J.Biol.Chem., 279, 2004
|
|
1UUQ
 
 | | Exo-mannosidase from Cellvibrio mixtus | | Descriptor: | GLYCEROL, MANNOSYL-OLIGOSACCHARIDE GLUCOSIDASE, SULFATE ION | | Authors: | Dias, M.V.F, Vincent, F, Pell, G, Prates, J.A.M, Centeno, M.S.J, Ferreira, L.M.A, Gilbert, H.J, Davies, G.J, Fontes, C.M.G.A. | | Deposit date: | 2004-01-09 | | Release date: | 2004-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights Into the Molecular Determinants of Substrate Specificity in Glycoside Hydrolase Family 5 Revealed by the Crystal Structure and Kinetics of Cellvibrio Mixtus Mannosidase 5A
J.Biol.Chem., 279, 2004
|
|
1GYE
 
 | | Structure of Cellvibrio cellulosa alpha-L-arabinanase complexed with Arabinohexaose | | Descriptor: | ARABINAN ENDO-1,5-ALPHA-L-ARABINOSIDASE A, CHLORIDE ION, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose | | Authors: | Nurizzo, D, Turkenburg, J.P, Charnock, S.J, Roberts, S.M, Dodson, E.J, McKie, V.A, Taylor, E.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cellvibrio japonicus alpha-L-arabinanase 43A has a novel five-blade beta-propeller fold.
Nat. Struct. Biol., 9, 2002
|
|
1GQJ
 
 | | Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with xylobiose | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-D-GLUCURONIDASE, COBALT (II) ION, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural Basis for Catalysis and Specificity of the Pseudomonas Cellulosa Alpha-Glucuronidase, Glca67A
Structure, 10, 2002
|
|
1GNY
 
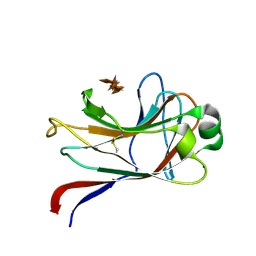 | | xylan-binding module CBM15 | | Descriptor: | SODIUM ION, XYLANASE 10C, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Szabo, S, Jamal, S, Xie, H, Charnock, S.J, Bolam, D.N, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of a Family 15 Carbohydrate-Binding Module in Complex with Xylopentaose: Evidence that Xylan Binds in an Approximate Three-Fold Helical Conformation
J.Biol.Chem., 276, 2001
|
|
1GQK
 
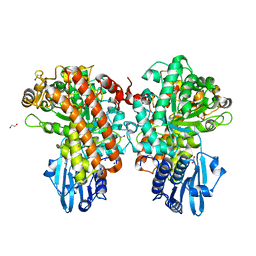 | | Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with glucuronic acid | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-D-GLUCURONIDASE, COBALT (II) ION, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural Basis for Catalysis and Specificity of the Pseudomonas Cellulosa Alpha-Glucuronidase, Glca67A
Structure, 10, 2002
|
|
1GQL
 
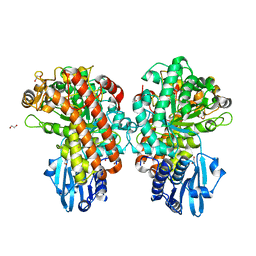 | | Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with glucuronic acid and xylotriose | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-D-GLUCURONIDASE, COBALT (II) ION, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Structural Basis for Catalysis and Specificity of the Pseudomonas Cellulosa Alpha-Glucuronidase, Glca67A
Structure, 10, 2002
|
|
1H41
 
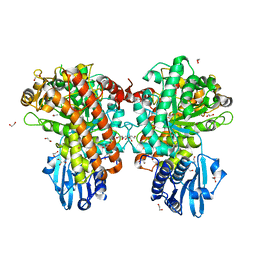 | | Pseudomonas cellulosa E292A alpha-D-glucuronidase mutant complexed with aldotriuronic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-alpha-D-glucopyranuronic acid, ALPHA-GLUCURONIDASE, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-09-25 | | Release date: | 2003-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Alpha-Glucuronidase,Glca67A,of Cellvibrio Japonicus Utilizes the Carboxylate and Methyl Groups of Aldobiouronic Acid as Important Substrate Recognition Determinants
J.Biol.Chem., 278, 2003
|
|
1HEJ
 
 | | C-terminal xylan binding domain from Cellulomonas fimi xylanase 11A | | Descriptor: | ENDO-1,4-BETA-XYLANASE D | | Authors: | Simpson, P.J, Hefang, X, Bolam, D.N, White, P, Hancock, S.M, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-10 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Evidence for Synergy between Family 2B Carbohydrate Binding Modules in Cellulomonas Fimi Xylanase 11A
Biochemistry, 40, 2001
|
|
1HEH
 
 | | C-terminal xylan binding domain from Cellulomonas fimi xylanase 11A | | Descriptor: | ENDO-1,4-BETA-XYLANASE D | | Authors: | Simpson, P.J, Hefang, X, Bolam, D.N, White, P, Hancock, S.M, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Evidence for Synergy between Family 2B Carbohydrate Binding Modules in Cellulomonas Fimi Xylanase 11A
Biochemistry, 40, 2001
|
|
1GYH
 
 | | Structure of D158A Cellvibrio cellulosa alpha-L-arabinanase mutant | | Descriptor: | ARABINAN ENDO-1,5-ALPHA-L-ARABINOSIDASE A, CHLORIDE ION | | Authors: | Nurizzo, D, Turkenburg, J.P, Charnock, S.J, Roberts, S.M, Dodson, E.J, McKie, V.A, Taylor, E.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Cellovibrio Cellulosa Alpha-L-Arabinanase 43A Has a Novel Five-Blade Beta-Propeller Fold
Nat.Struct.Biol., 9, 2002
|
|
1GYD
 
 | | Structure of Cellvibrio cellulosa alpha-L-arabinanase | | Descriptor: | ARABINAN ENDO-1,5-ALPHA-L-ARABINOSIDASE A | | Authors: | Nurizzo, D, Turkenburg, J.P, Charnock, S.J, Roberts, S.M, Dodson, E.J, McKie, V.A, Taylor, E.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cellvibrio japonicus alpha-L-arabinanase 43A has a novel five-blade beta-propeller fold.
Nat. Struct. Biol., 9, 2002
|
|
1GQI
 
 | | Structure of Pseudomonas cellulosa alpha-D-glucuronidase | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-GLUCURONIDASE, COBALT (II) ION, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The Structural Basis for Catalysis and Specificity of the Pseudomonas Cellulosa Alpha-Glucuronidase, Glca67A
Structure, 10, 2002
|
|
1J9Y
 
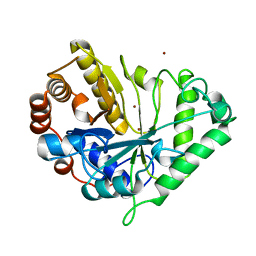 | | Crystal structure of mannanase 26A from Pseudomonas cellulosa | | Descriptor: | MANNANASE A, ZINC ION | | Authors: | Hogg, D, Woo, E.-J, Bolam, D.N, McKie, V.A, Gilbert, H.J, Pickersgill, R.W. | | Deposit date: | 2001-05-29 | | Release date: | 2001-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of mannanase 26A from Pseudomonas cellulosa and analysis of residues involved in substrate binding
J.Biol.Chem., 276, 2001
|
|
3W5M
 
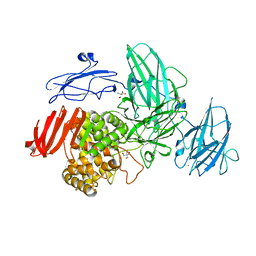 | | Crystal Structure of Streptomyces avermitilis alpha-L-rhamnosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Putative rhamnosidase | | Authors: | Fujimoto, Z, Jackson, A, Michikawa, M, Maehara, T, Momma, M, Henrissat, B.F, Gilbert, H.J, Kaneko, S. | | Deposit date: | 2013-01-31 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a Streptomyces avermitilis alpha-L-rhamnosidase reveals a novel carbohydrate-binding module CBM67 within the six-domain arrangement.
J.Biol.Chem., 288, 2013
|
|
3W5N
 
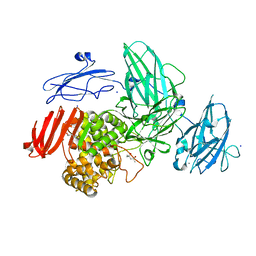 | | Crystal Structure of Streptomyces avermitilis alpha-L-rhamnosidase complexed with L-rhamnose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Putative rhamnosidase, ... | | Authors: | Fujimoto, Z, Jackson, A, Michikawa, M, Maehara, T, Momma, M, Henrissat, B.F, Gilbert, H.J, Kaneko, S. | | Deposit date: | 2013-01-31 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a Streptomyces avermitilis alpha-L-rhamnosidase reveals a novel carbohydrate-binding module CBM67 within the six-domain arrangement.
J.Biol.Chem., 288, 2013
|
|
3ZXJ
 
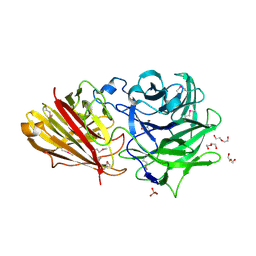 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, HIAXHD3, ... | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZXK
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIAXHD3, alpha-L-arabinofuranose-(1-2)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZXL
 
 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HIAXHD3 | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AFD
 
 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens with a partially bound cellotetraose moeity. | | Descriptor: | ENDOGLUCANASE CEL5A, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-18 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Understanding How Noncatalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan.
J.Biol.Chem., 288, 2013
|
|
4AFM
 
 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens. | | Descriptor: | ACETATE ION, ENDOGLUCANASE CEL5A, GLYCEROL | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan.
J.Biol.Chem., 288, 2013
|
|
