3BFQ
 
 | |
3BWU
 
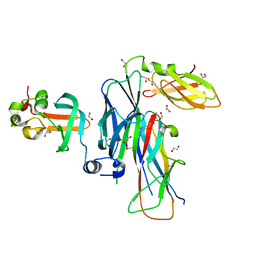 | | Crystal structure of the ternary complex of FimD (N-Terminal Domain, FimDN) with FimC and the N-terminally truncated pilus subunit FimF (FimFt) | | 分子名称: | 1,2-ETHANEDIOL, Chaperone protein fimC, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Eidam, O, Grutter, M.G, Capitani, G. | | 登録日 | 2008-01-10 | | 公開日 | 2008-03-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of the ternary FimC-FimF(t)-FimD(N) complex indicates conserved pilus chaperone-subunit complex recognition by the usher FimD
Febs Lett., 582, 2008
|
|
5IQN
 
 | | Crystal structure of the E. coli type 1 pilus subunit FimG (engineered variant with substitution Q134E; N-terminal FimG residues 1-12 truncated) in complex with the donor strand peptide DsF_SRIRIRGYVR | | 分子名称: | 1,2-ETHANEDIOL, COBALT (II) ION, Protein FimF, ... | | 著者 | Giese, C, Eras, J, Kern, A, Scharer, M.A, Capitani, G, Glockshuber, R. | | 登録日 | 2016-03-11 | | 公開日 | 2016-07-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Accelerating the Association of the Most Stable Protein-Ligand Complex by More than Two Orders of Magnitude.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5IQO
 
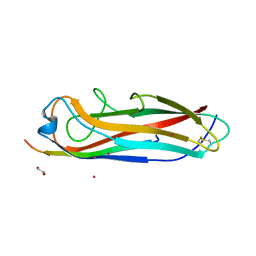 | | Crystal structure of the E. coli type 1 pilus subunit FimG (engineered variant with substitutions Q134E and S138E; N-terminal FimG residues 1-12 truncated) in complex with the donor strand peptide DsF_T4R-T6R-D13N | | 分子名称: | 1,2-ETHANEDIOL, COBALT (II) ION, PENTAETHYLENE GLYCOL, ... | | 著者 | Giese, C, Eras, J, Kern, A, Capitani, G, Glockshuber, R. | | 登録日 | 2016-03-11 | | 公開日 | 2016-07-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.302 Å) | | 主引用文献 | Accelerating the Association of the Most Stable Protein-Ligand Complex by More than Two Orders of Magnitude.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5IQM
 
 | | Crystal structure of the E. coli type 1 pilus subunit FimG (engineered variant with substitution Q134E; N-terminal FimG residues 1-12 truncated) in complex with the donor strand peptide DsF_T4R-T6R-D13N | | 分子名称: | COBALT (II) ION, Protein FimF, Protein FimG | | 著者 | Giese, C, Eras, J, Kern, A, Scharer, M.A, Capitani, G, Glockshuber, R. | | 登録日 | 2016-03-11 | | 公開日 | 2016-07-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Accelerating the Association of the Most Stable Protein-Ligand Complex by More than Two Orders of Magnitude.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5LB7
 
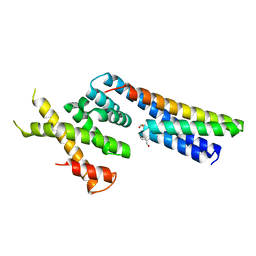 | | Complex structure between p60N/p80C katanin and a peptide derived from ASPM | | 分子名称: | Abnormal spindle-like microcephaly-associated protein homolog, Katanin p60 ATPase-containing subunit A1, Katanin p80 WD40 repeat-containing subunit B1 | | 著者 | Rezabkova, L, Capitani, G, Kammerer, R.A, Steinmetz, M.O. | | 登録日 | 2016-06-15 | | 公開日 | 2017-04-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex.
Nat. Cell Biol., 19, 2017
|
|
5LP9
 
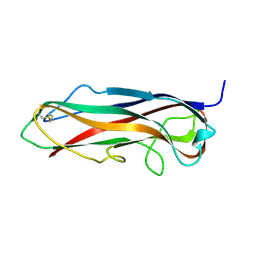 | | FimA wt from S. flexneri | | 分子名称: | Major type 1 subunit fimbrin (Pilin) | | 著者 | Zyla, D, Capitani, G, Prota, A, Glockshuber, R. | | 登録日 | 2016-08-12 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (0.88626635 Å) | | 主引用文献 | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 2019
|
|
2DGL
 
 | | Crystal structure of Escherichia coli GadB in complex with bromide | | 分子名称: | ACETIC ACID, BROMIDE ION, Glutamate decarboxylase beta, ... | | 著者 | Gruetter, M.G, Capitani, G, Gut, H. | | 登録日 | 2006-03-14 | | 公開日 | 2006-06-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
2DGM
 
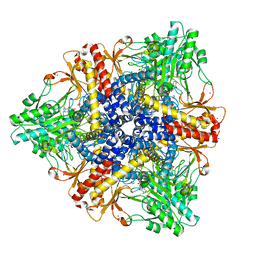 | | Crystal structure of Escherichia coli GadB in complex with iodide | | 分子名称: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | 著者 | Gruetter, M.G, Capitani, G, Gut, H. | | 登録日 | 2006-03-14 | | 公開日 | 2006-06-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
2DGK
 
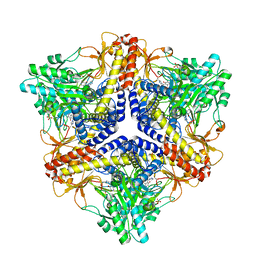 | | Crystal structure of an N-terminal deletion mutant of Escherichia coli GadB in an autoinhibited state (aldamine) | | 分子名称: | 1,2-ETHANEDIOL, Glutamate decarboxylase beta, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Gruetter, M.G, Capitani, G, Gut, H. | | 登録日 | 2006-03-14 | | 公開日 | 2006-06-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
5NBT
 
 | | Apo structure of p60N/p80C katanin | | 分子名称: | Katanin p60 ATPase-containing subunit A1, Katanin p80 WD40 repeat-containing subunit B1 | | 著者 | Jiang, K, Rezabkova, L, Hua, S, Liu, Q, Capitani, G, Altelaar, A.F.M, Heck, A.J.R, Kammerer, R.A, Steinmetz, M.O, Akhmanova, A. | | 登録日 | 2017-03-02 | | 公開日 | 2017-04-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex.
Nat. Cell Biol., 19, 2017
|
|
6ZRW
 
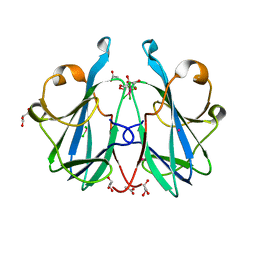 | | Crystal structure of the fungal lectin CML1 | | 分子名称: | ACETATE ION, GLYCEROL, Mucin-binding lectin 1, ... | | 著者 | Bleuler-Martinez, S, Olieric, V, Sharpe, M, Capitani, G, Aebi, M, Kuenzler, M. | | 登録日 | 2020-07-15 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-03-01 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structure-function relationship of a novel fucoside-binding fruiting body lectin from Coprinopsis cinerea exhibiting nematotoxic activity.
Glycobiology, 32, 2022
|
|
7B0W
 
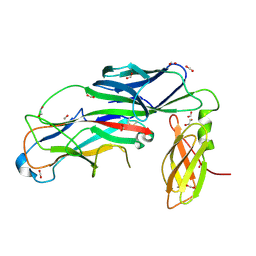 | | Crystal structure of the E. coli type 1 pilus assembly inhibitor FimI bound to FimC | | 分子名称: | 1,2-ETHANEDIOL, Chaperone protein FimC, FORMIC ACID, ... | | 著者 | Scharer, M.A, Zigova, Z, Giese, C, Puorger, C, Ignatov, O, Capitani, G, Glockshuber, R. | | 登録日 | 2020-11-23 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Comprehensive kinetic characterization of bacterial pilus rod assembly and assembly termination
To Be Published
|
|
7B0X
 
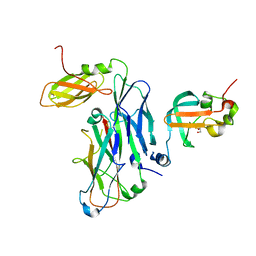 | | Crystal structure of the ternary complex of the E. coli type 1 pilus proteins FimC, FimI and the N-terminal domain of FimD | | 分子名称: | 1,2-ETHANEDIOL, Chaperone protein FimC, Fimbrin-like protein FimI, ... | | 著者 | Scharer, M.A, Zigova, Z, Giese, C, Puorger, C, Ignatov, O, Capitani, G, Glockshuber, R. | | 登録日 | 2020-11-23 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Comprehensive kinetic characterization of bacterial pilus rod assembly and assembly termination
To Be Published
|
|
5NKT
 
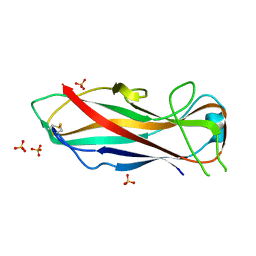 | | FimA wt from E. coli | | 分子名称: | SULFATE ION, Type-1 fimbrial protein, A chain | | 著者 | Zyla, D, Capitani, G, Prota, A, Glockshuber, R. | | 登録日 | 2017-04-03 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 2019
|
|
5OW5
 
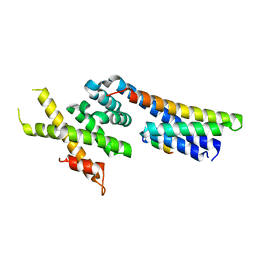 | | p60p80-CAMSAP complex | | 分子名称: | 1,2-ETHANEDIOL, Calmodulin-regulated spectrin-associated protein, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Rezabkova, L, Capitani, G, Prota, A.E, Kammerer, R.A, Steinmetz, M.O. | | 登録日 | 2017-08-30 | | 公開日 | 2018-07-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Basis of Formation of the Microtubule Minus-End-Regulating CAMSAP-Katanin Complex.
Structure, 26, 2018
|
|
3G7Y
 
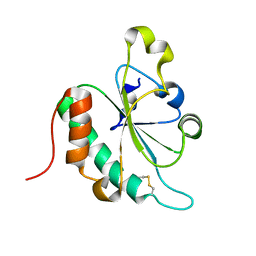 | | Crystal structure of oxidized Ost6L | | 分子名称: | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6 | | 著者 | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | 登録日 | 2009-02-11 | | 公開日 | 2009-06-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.215 Å) | | 主引用文献 | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GA4
 
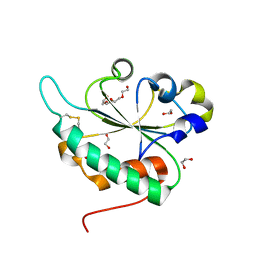 | | Crystal structure of Ost6L (photoreduced form) | | 分子名称: | 1,2-ETHANEDIOL, Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6, TETRAETHYLENE GLYCOL | | 著者 | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | 登録日 | 2009-02-16 | | 公開日 | 2009-06-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G9B
 
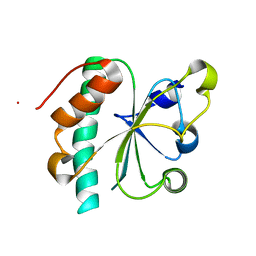 | | Crystal structure of reduced Ost6L | | 分子名称: | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6 | | 著者 | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | 登録日 | 2009-02-13 | | 公開日 | 2009-06-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HBX
 
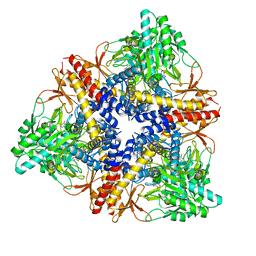 | | Crystal structure of GAD1 from Arabidopsis thaliana | | 分子名称: | Glutamate decarboxylase 1 | | 著者 | Gut, H, Dominici, P, Pilati, S, Gruetter, M.G, Capitani, G. | | 登録日 | 2009-05-05 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.672 Å) | | 主引用文献 | A common structural basis for pH- and calmodulin-mediated regulation in plant glutamate decarboxylase.
J.Mol.Biol., 392, 2009
|
|
3MAU
 
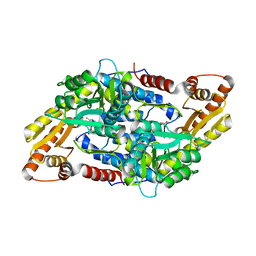 | | Crystal structure of StSPL in complex with phosphoethanolamine | | 分子名称: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, PHOSPHATE ION, sphingosine-1-phosphate lyase, ... | | 著者 | Bourquin, F, Grutter, M.G, Capitani, G. | | 登録日 | 2010-03-24 | | 公開日 | 2010-08-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure and Function of Sphingosine-1-Phosphate Lyase, a Key Enzyme of Sphingolipid Metabolism.
Structure, 18, 2010
|
|
3MAD
 
 | |
3MBB
 
 | | Crystal structure of StSPL - apo form, after treatment with semicarbazide | | 分子名称: | 1,2-ETHANEDIOL, PHOSPHATE ION, Putative sphingosine-1-phosphate lyase, ... | | 著者 | Bourquin, F, Grutter, M.G, Capitani, G. | | 登録日 | 2010-03-25 | | 公開日 | 2010-08-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.051 Å) | | 主引用文献 | Structure and Function of Sphingosine-1-Phosphate Lyase, a Key Enzyme of Sphingolipid Metabolism.
Structure, 18, 2010
|
|
3MC6
 
 | | Crystal structure of ScDPL1 | | 分子名称: | PHOSPHATE ION, Sphingosine-1-phosphate lyase | | 著者 | Bourquin, F, Grutter, M.G, Capitani, G. | | 登録日 | 2010-03-27 | | 公開日 | 2010-08-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structure and Function of Sphingosine-1-Phosphate Lyase, a Key Enzyme of Sphingolipid Metabolism.
Structure, 18, 2010
|
|
3MAF
 
 | | Crystal structure of StSPL (asymmetric form) | | 分子名称: | CHLORIDE ION, PHOSPHATE ION, sphingosine-1-phosphate lyase | | 著者 | Bourquin, F, Grutter, M.G, Capitani, G. | | 登録日 | 2010-03-23 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.971 Å) | | 主引用文献 | Structure and Function of Sphingosine-1-Phosphate Lyase, a Key Enzyme of Sphingolipid Metabolism.
Structure, 18, 2010
|
|
