6NPC
 
 | | X-ray crystal structure of TmpA, 2-trimethylaminoethylphosphonate hydroxylase, with Fe, 2OG, and 2-trimethylaminoethylphosphonate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, N,N,N-trimethyl-2-phosphonoethan-1-aminium, ... | | Authors: | Rajakovich, L.J, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases.
Biochemistry, 58, 2019
|
|
6NPD
 
 | | X-ray crystal structure of TmpA, 2-trimethylaminoethylphosphonate hydroxylase, with Fe, 2OG, and (R)-1-hydroxy-2-trimethylaminoethylphosphonate | | Descriptor: | (2R)-2-hydroxy-N,N,N-trimethyl-2-phosphonoethan-1-aminium, FE (II) ION, TmpA, ... | | Authors: | Rajakovich, L.J, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases.
Biochemistry, 58, 2019
|
|
6NPB
 
 | | X-ray crystal structure of TmpA, 2-trimethylaminoethylphosphonate hydroxylase, with Fe and 2OG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, SULFATE ION, ... | | Authors: | Rajakovich, L.J, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases.
Biochemistry, 58, 2019
|
|
6VP5
 
 | | Ethylene forming enzyme (EFE) D191E variant in complex with Fe(II), L-arginine, and 2OG | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ... | | Authors: | Davis, K.M, Copeland, R.A, Boal, A.K. | | Deposit date: | 2020-02-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | An Iron(IV)-Oxo Intermediate Initiating l-Arginine Oxidation but Not Ethylene Production by the 2-Oxoglutarate-Dependent Oxygenase, Ethylene-Forming Enzyme.
J.Am.Chem.Soc., 143, 2021
|
|
6VZY
 
 | |
6VP4
 
 | | Ethylene forming enzyme (EFE) in complex with Fe(II), L-arginine, and 2OG | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ... | | Authors: | Davis, K.M, Copeland, R.A, Boal, A.K. | | Deposit date: | 2020-02-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | An Iron(IV)-Oxo Intermediate Initiating l-Arginine Oxidation but Not Ethylene Production by the 2-Oxoglutarate-Dependent Oxygenase, Ethylene-Forming Enzyme.
J.Am.Chem.Soc., 143, 2021
|
|
6XCV
 
 | |
6ALO
 
 | | VioC L-arginine hydroxylase bound to Fe(II), L-arginine, and a peroxysuccinate intermediate | | Descriptor: | 4-peroxy-4-oxobutanoic acid, ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | Authors: | Dunham, N.P, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase.
J. Am. Chem. Soc., 139, 2017
|
|
6ALN
 
 | | VioC L-arginine hydroxylase bound to Fe(II), 3S-hydroxy-L-arginine, and 2OG | | Descriptor: | (2S,3S)-3-HYDROXYARGININE, 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | Authors: | Mitchell, A.J, Dunham, N.P, Boal, A.K. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase.
J. Am. Chem. Soc., 139, 2017
|
|
6ALM
 
 | | VioC L-arginine hydroxylase bound to Fe(II), L-arginine, and 2-OXO-GLUTARIC ACID | | Descriptor: | 2-OXOGLUTARIC ACID, ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | Authors: | Dunham, N.P, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase.
J. Am. Chem. Soc., 139, 2017
|
|
6ALQ
 
 | | VioC L-arginine hydroxylase bound to Fe(II), L-arginine, and succinate | | Descriptor: | ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, FE (II) ION, ... | | Authors: | Dunham, N.P, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase.
J. Am. Chem. Soc., 139, 2017
|
|
6ALR
 
 | | VioC L-arginine hydroxylase bound to the vanadyl ion, L-arginine, and succinate | | Descriptor: | ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, SUCCINIC ACID, ... | | Authors: | Mitchell, A.J, Dunham, N.P, Bergman, J.A, Boal, A.K. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase.
J. Am. Chem. Soc., 139, 2017
|
|
6ALP
 
 | | VioC L-arginine hydroxylase bound to Fe(II), 3S-hydroxy-L-arginine, and succinate | | Descriptor: | (2S,3S)-3-HYDROXYARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, FE (II) ION, ... | | Authors: | Mitchell, A.J, Dunham, N.P, Boal, A.K. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Visualizing the Reaction Cycle in an Iron(II)- and 2-(Oxo)-glutarate-Dependent Hydroxylase.
J. Am. Chem. Soc., 139, 2017
|
|
4N6W
 
 | | X-Ray Crystal Structure of Citrate-bound PhnZ | | Descriptor: | CITRATE ANION, FE (III) ION, Predicted HD phosphohydrolase PhnZ | | Authors: | Worsdorfer, B, Lingaraju, M, Yennawar, N.H, Boal, A.K, Krebs, C, Bollinger Jr, J.M, Pandelia, M.E. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Organophosphonate-degrading PhnZ reveals an emerging family of HD domain mixed-valent diiron oxygenases.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4N71
 
 | | X-Ray Crystal Structure of 2-amino-1-hydroxyethylphosphonate-bound PhnZ | | Descriptor: | FE (III) ION, Predicted HD phosphohydrolase PhnZ, [(1R)-2-amino-1-hydroxyethyl]phosphonic acid | | Authors: | Woersdoerfer, B, Lingaraju, M, Yennawar, N, Boal, A.K, Krebs, C, Bollinger Jr, J.M, Pandelia, M.-E. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | Organophosphonate-degrading PhnZ reveals an emerging family of HD domain mixed-valent diiron oxygenases.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8CVB
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Structure of the H6H cyclization reactant complex
To Be Published
|
|
8CVF
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Hyoscyamine 6-beta-hydroxylase, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Structure of the L289F H6H ferryl-mimicking complex
To Be Published
|
|
8CVH
 
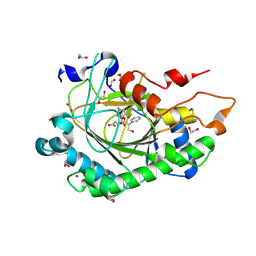 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the L289F H6H cyclization ferryl-mimicking complex
To Be Published
|
|
8CVD
 
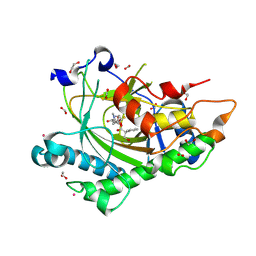 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, succinate, and scopolamine | | Descriptor: | (1R,2R,4S,5S,7s)-9-methyl-3-oxa-9-azatricyclo[3.3.1.0~2,4~]nonan-7-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FE (II) ION, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Structure of the H6H cyclization product complex
To Be Published
|
|
8CVC
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structure mimicking the H6H cyclization ferryl complex
To Be Published
|
|
8CVG
 
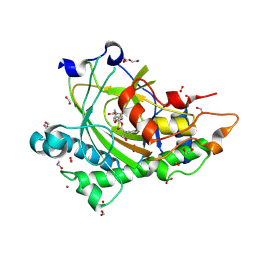 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of the L289F H6H cyclization reactant complex
To Be Published
|
|
8CVA
 
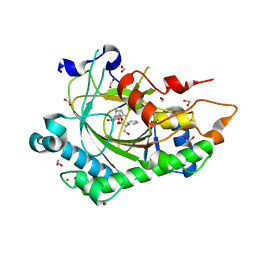 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, succinate, and 6-OH-hyoscyamine | | Descriptor: | (1R,3S,5R,6S)-6-hydroxy-8-methyl-8-azabicyclo[3.2.1]octan-3-yl (2S)-3-hydroxy-2-phenylpropanoate, 1,2-ETHANEDIOL, FE (II) ION, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Structure of the H6H hydroxylation product complex
To Be Published
|
|
8CV8
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Structure of the H6H hydroxylation reactant complex
To Be Published
|
|
8CV9
 
 | | Structure of Hyoscyamine 6-beta Hydroxylase in complex with vanadyl, succinate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Hyoscyamine 6-beta-hydroxylase, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of the H6H hydroxylation reactant complex
To Be Published
|
|
8CVE
 
 | | Structure of L289F Hyoscyamine 6-beta Hydroxylase in complex with iron, 2-oxoglutarate, and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wenger, E.W, Boal, A.K, Bollinger, J.M, Krebs, C. | | Deposit date: | 2022-05-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure of the L289F H6H hydroxylation reactant complex
To Be Published
|
|
