6MS3
 
 | | Crystal structure of the GH43 protein BlXynB mutant (K247S) from Bacillus licheniformis | | Descriptor: | CALCIUM ION, GLYCEROL, Glycoside Hydrolase Family 43, ... | | Authors: | Zanphorlin, L.M, Morais, M.A.B, Diogo, J.A, Murakami, M.T. | | Deposit date: | 2018-10-16 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design combined with evolutionary diversity led to the discovery of the xylose-releasing exo-xylanase activity in the glycoside hydrolase family 43.
Biotechnol. Bioeng., 116, 2019
|
|
6MS2
 
 | | Crystal structure of the GH43 BlXynB protein from Bacillus licheniformis | | Descriptor: | CALCIUM ION, Glycoside Hydrolase Family 43 | | Authors: | Zanphorlin, L.M, Morais, M.A.B, Diogo, J.A, Murakami, M.T. | | Deposit date: | 2018-10-16 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Structure-guided design combined with evolutionary diversity led to the discovery of the xylose-releasing exo-xylanase activity in the glycoside hydrolase family 43.
Biotechnol. Bioeng., 116, 2019
|
|
5DT5
 
 | | Crystal structure of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7 in space group P21 | | Descriptor: | Beta-glucosidase, SULFATE ION | | Authors: | Zanphorlin, L.M, Giuseppe, P.O, Tonoli, C.C.C, Murakami, M.T. | | Deposit date: | 2015-09-17 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Oligomerization as a strategy for cold adaptation: Structure and dynamics of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7.
Sci Rep, 6, 2016
|
|
5DT7
 
 | | Crystal structure of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7 in space group C2221 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Zanphorlin, L.M, Giuseppe, P.O, Tonoli, C.C.C, Murakami, M.T. | | Deposit date: | 2015-09-17 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Oligomerization as a strategy for cold adaptation: Structure and dynamics of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7.
Sci Rep, 6, 2016
|
|
6WIU
 
 | |
8D8P
 
 | |
6UNV
 
 | |
5WKA
 
 | | Crystal structure of a GH1 beta-glucosidase retrieved from microbial metagenome of Poraque Amazon lake | | Descriptor: | Beta-glucosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Morais, M.A.B, Toyama, D, Ramos, F.C, Zanphorlin, L.M, Tonoli, C.C.C, Miranda, F.P, Ruller, R, Henrique-Silva, F, Murakami, M.T. | | Deposit date: | 2017-07-24 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A novel beta-glucosidase isolated from the microbial metagenome of Lake Poraque (Amazon, Brazil).
Biochim. Biophys. Acta, 1866, 2018
|
|
4MDP
 
 | | Crystal structure of a GH1 beta-glucosidase from the fungus Humicola insolens in complex with glucose | | Descriptor: | Beta-glucosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Giuseppe, P.O, Souza, T.A.C.B, Souza, F.H.M, Zanphorlin, L.M, Machado, C.B, Ward, R.J, Jorge, J.A, Furriel, R.P.M, Murakami, M.T. | | Deposit date: | 2013-08-23 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for glucose tolerance in GH1 beta-glucosidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MDO
 
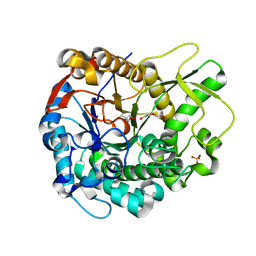 | | Crystal structure of a GH1 beta-glucosidase from the fungus Humicola insolens | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Giuseppe, P.O, Souza, T.A.C.B, Souza, F.H.M, Zanphorlin, L.M, Machado, C.B, Ward, R.J, Jorge, J.A, Furriel, R.P.M, Murakami, M.T. | | Deposit date: | 2013-08-23 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for glucose tolerance in GH1 beta-glucosidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6N99
 
 | | Xylose isomerase 2F1 variant from Streptomyces sp. F-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Miyamoto, R.Y, Vieira, P.S, Murakami, M.T, Zanphorlin, L.M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a novel xylose isomerase from Streptomyces sp. F-1 revealed the presence of unique features that differ from conventional classes.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6N98
 
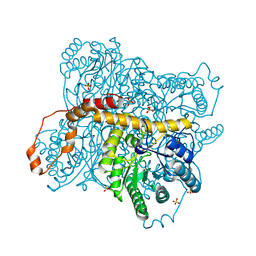 | | Xylose isomerase 1F1 variant from Streptomyces sp. F-1 | | Descriptor: | MAGNESIUM ION, SULFATE ION, Xylose isomerase | | Authors: | Miyamoto, R.Y, Vieira, P.S, Murakami, M.T, Zanphorlin, L.M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a novel xylose isomerase from Streptomyces sp. F-1 revealed the presence of unique features that differ from conventional classes.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
5BWF
 
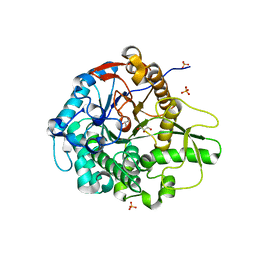 | | Crystal structure of the beta-glucosidase from Trichoderma harzianum | | Descriptor: | Beta-1,4-glucosidase, GLYCEROL, SULFATE ION | | Authors: | Santos, C.A, Zanphorlin, L.M, Crucello, A, Tonoli, C.C.C, Ruller, R, Souza, A.P, Murakami, M.T. | | Deposit date: | 2015-06-07 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the beta-glucosidase from Trichoderma harzianum
To Be Published
|
|
5C2Z
 
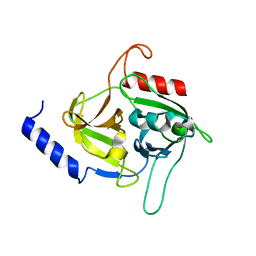 | | Molecular insights into the specificity of exfoliative toxins from Staphylococcus aureus | | Descriptor: | Exfoliative toxin D2 | | Authors: | Mariutti, R.B, Souza, T.A.C.B, Ullah, A, Zanphorlin, L.M, Murakami, M.T, Arni, R.K. | | Deposit date: | 2015-06-16 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9553 Å) | | Cite: | Crystal structure of Staphylococcus aureus exfoliative toxin D-like protein: Structural basis for the high specificity of exfoliative toxins.
Biochem.Biophys.Res.Commun., 467, 2015
|
|
4PN2
 
 | | Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylotriose | | Descriptor: | CALCIUM ION, Xylanase, beta-D-xylopyranose | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
4PMZ
 
 | | Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylobiose | | Descriptor: | CALCIUM ION, Xylanase, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
5CZL
 
 | | Crystal structure of a novel GH8 endo-beta-1,4-glucanase from an Achatina fulica gut metagenomic library | | Descriptor: | Glucanase, PHOSPHATE ION | | Authors: | Scapin, S.M.N, Souza, F.H.M, Zanphorlin, L.M, Almeida, T.S, Sade, Y.B, Cardoso, A.M, Pinheiro, G.L, Murakami, M.T. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Crystal structure of a novel GH8 endo-beta-1,4-glucanase from an Achatina fulica gut metagenomic library
To Be Published
|
|
4PMV
 
 | | Crystal structure of a novel reducing-end xylose-releasing exo-oligoxylanase (XynA) belonging to GH10 family (space group P43212) | | Descriptor: | Endo-1,4-beta-xylanase A | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
4PMU
 
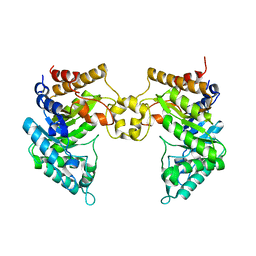 | | Crystal structure of a novel reducing-end xylose-releasing exo-oligoxylanase (XynA) belonging to GH10 family (space group P1211) | | Descriptor: | Endo-1,4-beta-xylanase A | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.857 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
4PMY
 
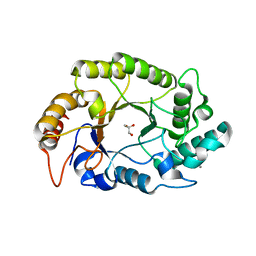 | | Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylose | | Descriptor: | CALCIUM ION, GLYCEROL, Xylanase, ... | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
4PMX
 
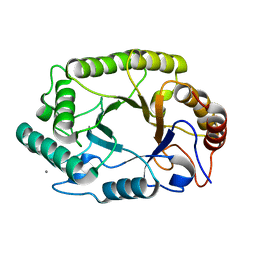 | | Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri in the native form | | Descriptor: | CALCIUM ION, Xylanase | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
4GSO
 
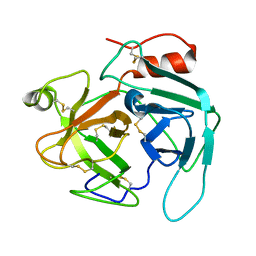 | | structure of Jararacussin-I | | Descriptor: | Thrombin-like enzyme BjussuSP-1 | | Authors: | Ullah, A, Souza, T.C.A.B, Zanphorlin, L.M, Mariutti, R, Sanata, S.V, Murakami, M.T, Arni, R.K. | | Deposit date: | 2012-08-28 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Jararacussin-I: The highly negatively charged catalytic interface contributes to macromolecular selectivity in snake venom thrombin-like enzymes.
Protein Sci., 22, 2013
|
|
6EFU
 
 | | Crystal structure of the double mutant L167W / P172L of the beta-glucosidase from Trichoderma harzianum | | Descriptor: | Beta-glucosidase, NITRATE ION | | Authors: | Morais, M.A.B, Santos, C.A, Tonoli, C.C.C, Souza, A.P, Murakami, M.T. | | Deposit date: | 2018-08-17 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An engineered GH1 beta-glucosidase displays enhanced glucose tolerance and increased sugar release from lignocellulosic materials.
Sci Rep, 9, 2019
|
|
6D25
 
 | | Crystal structure of the GH51 arabinofuranosidase from Xanthomonas axonopodis pv. citri | | Descriptor: | Alpha-L-arabinosidase, GLYCEROL | | Authors: | Santos, C.R, Morais, M.A.B, Tonoli, C.C.C, Giuseppe, P.O, Murakami, M.T. | | Deposit date: | 2018-04-13 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The mechanism by which a distinguishing arabinofuranosidase can cope with internal di-substitutions in arabinoxylans.
Biotechnol Biofuels, 11, 2018
|
|
5KLE
 
 | | Structure of CBM_E1, a novel carbohydrate-binding module found by sugar cane soil metagenome, complexed with cellopentaose | | Descriptor: | Carbohydrate binding module E1, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Liberato, M.V, Campos, B.M, Zeri, A.C.M, Squina, F.M. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Novel Carbohydrate-binding Module from Sugar Cane Soil Metagenome Featuring Unique Structural and Carbohydrate Affinity Properties.
J.Biol.Chem., 291, 2016
|
|
