2WZM
 
 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-11-30 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
2WZT
 
 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-12-03 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
3WCY
 
 | |
2GRV
 
 | | Crystal Structure of LpqW | | Descriptor: | LpqW | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hijacking of a Substrate-binding Protein Scaffold for use in Mycobacterial Cell Wall Biosynthesis
J.Mol.Biol., 359, 2006
|
|
2ZZ8
 
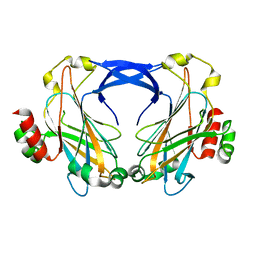 | |
3CKQ
 
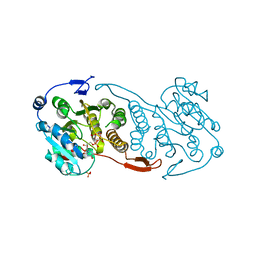 | | Crystal Structure of a Mycobacterial Protein | | Descriptor: | MANGANESE (II) ION, Putative uncharacterized protein, SULFATE ION, ... | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2008-03-16 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species.
J.Biol.Chem., 283, 2008
|
|
3CKN
 
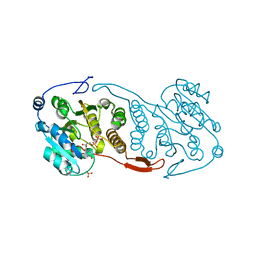 | | Crystal Structure of a Mycobacterial Protein | | Descriptor: | MANGANESE (II) ION, Putative uncharacterized protein, SULFATE ION, ... | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2008-03-16 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species.
J.Biol.Chem., 283, 2008
|
|
3CKV
 
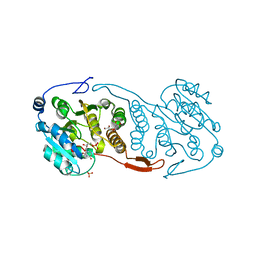 | | Crystal Structure of a Mycobacterial Protein | | Descriptor: | GLYCEROL, Putative uncharacterized protein, SULFATE ION, ... | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species.
J.Biol.Chem., 283, 2008
|
|
3CKO
 
 | | Crystal Structure of a Mycobacterial Protein | | Descriptor: | GLYCEROL, Putative uncharacterized protein, SULFATE ION, ... | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2008-03-16 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species.
J.Biol.Chem., 283, 2008
|
|
3CKJ
 
 | | Crystal Structure of a Mycobacterial Protein | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CITRIC ACID, PHOSPHATE ION, ... | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2008-03-15 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species.
J.Biol.Chem., 283, 2008
|
|
5N29
 
 | |
