4H9V
 
 | | Structure of Geobacillus kaustophilus lactonase, mutant E101G/R230C with Zn2+ | | Descriptor: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4H9T
 
 | | Structure of Geobacillus kaustophilus lactonase, mutant E101N with bound N-butyryl-DL-homoserine lactone | | Descriptor: | FE (III) ION, MANGANESE (II) ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-24 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4H9Y
 
 | | Structure of Geobacillus kaustophilus lactonase, mutant E101N with Zn2+ | | Descriptor: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4H9Z
 
 | | Structure of Geobacillus kaustophilus lactonase, mutant E101N with Mn2+ | | Descriptor: | FE (III) ION, MANGANESE (II) ION, Phosphotriesterase | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4H9U
 
 | | Structure of Geobacillus kaustophilus lactonase, wild-type with Zn2+ | | Descriptor: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4HA0
 
 | | Structure of Geobacillus kaustophilus lactonase, mutant R230D with Zn2+ | | Descriptor: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4H9X
 
 | | Structure of Geobacillus kaustophilus lactonase, mutant E101G/R230C/D266N with Zn2+ and bound N-butyryl-DL-homoserine lactone | | Descriptor: | FE (III) ION, HYDROXIDE ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, ... | | Authors: | Xue, B, Chow, J.Y, Yew, W.S, Robinson, R.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-07 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural evidence of a productive active site architecture for an evolved quorum-quenching GKL lactonase.
Biochemistry, 52, 2013
|
|
4PL8
 
 | |
4PL7
 
 | |
8YJO
 
 | | Structure of E. coli glycyl radical enzyme PflD with bound malonate | | Descriptor: | MALONATE ION, Probable dehydratase PflD | | Authors: | Xue, B, Wei, Y, Robinson, R.C, Yew, W.S, Zhang, Y. | | Deposit date: | 2024-03-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
8YJN
 
 | | Structure of E. coli glycyl radical enzyme YbiW with bound glycerol | | Descriptor: | GLYCEROL, Probable dehydratase YbiW | | Authors: | Xue, B, Wei, Y, Robinson, R.C, Yew, W.S, Zhang, Y. | | Deposit date: | 2024-03-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
2FF3
 
 | | Crystal structure of Gelsolin domain 1:N-wasp V2 motif hybrid in complex with actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Xue, B, Aguda, A.H, Robinson, R.C. | | Deposit date: | 2005-12-19 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structural Basis of Actin Interaction with Multiple WH2/beta-Thymosin Motif-Containing Proteins
Structure, 14, 2006
|
|
4PH6
 
 | |
3OJG
 
 | | Structure of an inactive lactonase from Geobacillus kaustophilus with bound N-butyryl-DL-homoserine lactone | | Descriptor: | FE (III) ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Tung, A, Robinson, R.C. | | Deposit date: | 2010-08-22 | | Release date: | 2010-10-27 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Directed evolution of a thermostable quorum-quenching lactonase from the amidohydrolase superfamily
J.Biol.Chem., 285, 2010
|
|
7V6D
 
 | | Structure of lipase B from Lasiodiplodia theobromae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Lipase B | | Authors: | Xue, B, Zhang, H.F, Nguyen, G.K.T, Yew, W.S. | | Deposit date: | 2021-08-20 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Lipase from Lasiodiplodia theobromae Efficiently Hydrolyses C8-C10 Methyl Esters for the Preparation of Medium-Chain Triglycerides' Precursors.
Int J Mol Sci, 22, 2021
|
|
6JSS
 
 | | Structure of Geobacillus kaustophilus lactonase, Y99P mutant | | Descriptor: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | Authors: | Xue, B, Yew, W.S. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Directed Computational Evolution of Quorum-Quenching Lactonases from the Amidohydrolase Superfamily.
Structure, 28, 2020
|
|
6JSU
 
 | |
6JST
 
 | |
7FHE
 
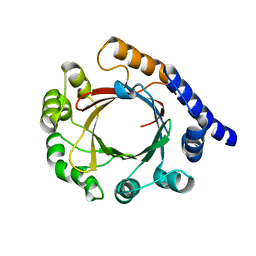 | | Structure of prenyltransferase mutant Q295F from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHC
 
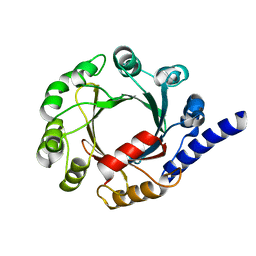 | | Structure of prenyltransferase mutant V49W from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHB
 
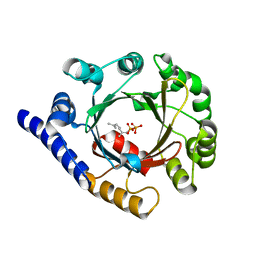 | | Structure of prenyltransferase from Streptomyces sp. (strain CL190) with bound GPP | | Descriptor: | GERANYL DIPHOSPHATE, Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHD
 
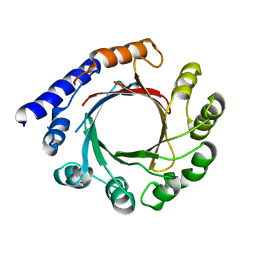 | | Structure of prenyltransferase mutant Y288P from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHF
 
 | | Structure of prenyltransferase mutant V49W/Y288F/Q295F from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7DHQ
 
 | | Structure of Halothiobacillus neapolitanus Microcompartments Protein CsoS1D | | Descriptor: | Microcompartments protein | | Authors: | Xue, B, Tan, Y.Q, Ali, S, Robinson, R.C, Narita, A, Yew, W.S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a Minimal alpha-Carboxysome-Derived Shell and Its Utility in Enzyme Stabilization.
Biomacromolecules, 22, 2021
|
|
5B6I
 
 | | Structure of fluorinase from Streptomyces sp. MA37 | | Descriptor: | ADENOSINE, Fluorinase, METHIONINE | | Authors: | Xue, B, Robinson, R.C. | | Deposit date: | 2016-05-29 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Directed Evolution of a Fluorinase for Improved Fluorination Efficiency with a Non-native Substrate
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
