1FI2
 
 | | CRYSTAL STRUCTURE OF GERMIN (OXALATE OXIDASE) | | Descriptor: | MANGANESE (II) ION, OXALATE OXIDASE | | Authors: | Woo, E.J, Dunwell, J.M, Goodenough, P.W, Marvier, A.C, Pickersgill, R.W. | | Deposit date: | 2000-08-03 | | Release date: | 2001-05-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities.
Nat.Struct.Biol., 7, 2000
|
|
1LRH
 
 | | Crystal structure of auxin-binding protein 1 in complex with 1-naphthalene acetic acid | | Descriptor: | NAPHTHALEN-1-YL-ACETIC ACID, ZINC ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Woo, E.J, Marshall, J, Bauly, J, Chen, J.-G, Venis, M, Napier, R.M, Pickersgill, R.W. | | Deposit date: | 2002-05-15 | | Release date: | 2002-06-19 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of auxin-binding protein 1 in complex with auxin.
EMBO J., 21, 2002
|
|
1LR5
 
 | | Crystal structure of auxin binding protein | | Descriptor: | Auxin binding protein 1, ZINC ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Woo, E.J, Marshall, J, Bauley, J, Chen, J.-G, Venis, M, Napier, R.M, Pickersgill, R.W. | | Deposit date: | 2002-05-14 | | Release date: | 2002-06-19 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of auxin-binding protein 1 in complex with auxin.
EMBO J., 21, 2002
|
|
6IY8
 
 | | DmpR-phenol complex of Pseudomonas putida | | Descriptor: | PHENOL, Positive regulator CapR, ZINC ION | | Authors: | Park, K.H, Woo, E.J. | | Deposit date: | 2018-12-13 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Tetrameric architecture of an active phenol-bound form of the AAA+transcriptional regulator DmpR.
Nat Commun, 11, 2020
|
|
6LDN
 
 | | Crystal structure of T.onnurineus Csm5 | | Descriptor: | Csm5 | | Authors: | Park, K.H, Woo, E.J. | | Deposit date: | 2019-11-22 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Csm5 subunit of the Type III-A Csm complex at 2.6 Angstroms resolution
To Be Published
|
|
6AIL
 
 | | CRYSTAL STRUCTURE AT 1.3 ANGSTROMS RESOLUTION OF A NOVEL UDG, UdgX, FROM Mycobacterium smegmatis | | Descriptor: | IRON/SULFUR CLUSTER, Uracil DNA glycosylase X | | Authors: | Ahn, W.C, Aroli, S, Varshney, V, Woo, E.J. | | Deposit date: | 2018-08-24 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.335 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
6AJR
 
 | | Complex form of Uracil DNA glycosylase X and uracil | | Descriptor: | IRON/SULFUR CLUSTER, URACIL, Uracil DNA glycosylase superfamily protein | | Authors: | Ahn, W.C, Aroli, S, Varshney, U, Woo, E.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.341 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
6AJO
 
 | | Complex form of Uracil DNA glycosylase X and uracil-DNA. | | Descriptor: | DNA (5'-D(P*(ORP)P*TP*T)-3'), IRON/SULFUR CLUSTER, PHOSPHATE ION, ... | | Authors: | Ahn, W.C, Aroli, S, Varshney, U, Woo, E.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
6AJP
 
 | | Complex form of Uracil DNA glycosylase X and deoxyuridine monophosphate. | | Descriptor: | 2'-DEOXYURIDINE-5'-MONOPHOSPHATE, IRON/SULFUR CLUSTER, Uracil DNA glycosylase superfamily protein | | Authors: | Ahn, W.C, Aroli, S, Varshney, U, Woo, E.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.334 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
6AJQ
 
 | | E52Q mutant form of Uracil DNA glycosylase X from Mycobacterium smegmatis. | | Descriptor: | IRON/SULFUR CLUSTER, Uracil DNA glycosylase superfamily protein | | Authors: | Ahn, W.C, Aroli, S, Varshney, U, Woo, E.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.342 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
6AJS
 
 | | H109S mutant form of Uracil DNA glycosylase X. | | Descriptor: | IRON/SULFUR CLUSTER, Uracil DNA glycosylase superfamily protein | | Authors: | Ahn, W.C, Aroli, S, Varshney, U, Woo, E.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
8IM8
 
 | | Crystal structure of Periplasmic alpha-amylase (MalS) from E.coli | | Descriptor: | CALCIUM ION, Periplasmic alpha-amylase | | Authors: | An, Y, Park, J.T, Park, K.H, Woo, E.J. | | Deposit date: | 2023-03-06 | | Release date: | 2023-05-24 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Distinctive Permutated Domain Structure of Periplasmic alpha-Amylase (MalS) from Glycoside Hydrolase Family 13 Subfamily 19.
Molecules, 28, 2023
|
|
3IQC
 
 | |
6M3T
 
 | |
6M3F
 
 | |
6M3U
 
 | |
6LYF
 
 | |
3K1H
 
 | |
4E1Q
 
 | | Crystal structure of Wheat Cyclophilin A at 1.25 A resolution | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Sekhon, S.S, Jeong, D.G, Woo, E.J, Singh, P, Yoon, T.S. | | Deposit date: | 2012-03-06 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Structural and biochemical characterization of the cytosolic wheat cyclophilin TaCypA-1
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4AEE
 
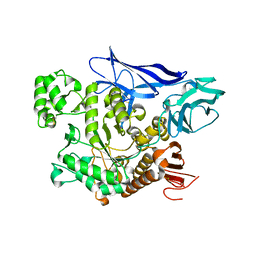 | | CRYSTAL STRUCTURE OF MALTOGENIC AMYLASE FROM S.MARINUS | | Descriptor: | ALPHA AMYLASE, CATALYTIC REGION | | Authors: | Jung, T.Y, Park, C.H, Yoon, S.M, Park, S.H, Park, K.H, Woo, E.J. | | Deposit date: | 2012-01-10 | | Release date: | 2012-01-18 | | Last modified: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Association of Novel Domain in Active Site of Archaic Hyperthermophilic Maltogenic Amylase from Staphylothermus Marinus.
J.Biol.Chem., 287, 2012
|
|
4HY7
 
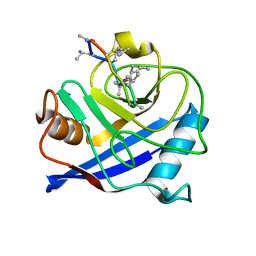 | | Structural and biochemical characterization of a cytosolic wheat cyclophilin TaCypA-1 | | Descriptor: | Cyclosporin A, Peptidyl-prolyl cis-trans isomerase | | Authors: | Sekhon, S.S, Jeong, D.G, Woo, E.J, Singh, P, Pareek, A, Yoon, T.-S. | | Deposit date: | 2012-11-13 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and biochemical characterization of the cytosolic wheat cyclophilin TaCypA-1.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5FRZ
 
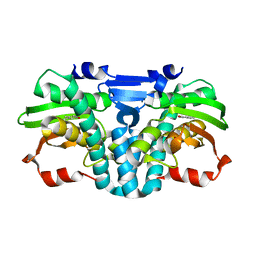 | |
5FRV
 
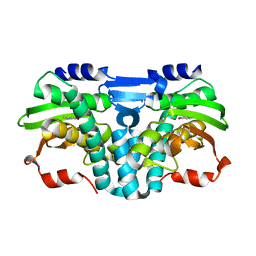 | |
5FRY
 
 | |
5FRX
 
 | |
