1WCU
 
 | | CBM29_1, A Family 29 Carbohydrate Binding Module from Piromyces equi | | Descriptor: | GLYCEROL, NON-CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davis, G.J, Gilbert, H.J. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W8U
 
 | | CBM29-2 mutant D83A complexed with mannohexaose: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-09-28 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W8T
 
 | | CBM29-2 mutant K74A complexed with cellulohexaose: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-09-28 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W8W
 
 | | CBM29-2 mutant Y46A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON-CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W8Z
 
 | | CBM29-2 mutant K85A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-10-01 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W90
 
 | | CBM29-2 mutant D114A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | 1,2-ETHANEDIOL, NON-CATALYTIC PROTEIN 1, SODIUM ION | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-10-01 | | Release date: | 2005-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W9F
 
 | | CBM29-2 mutant R112A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-10-12 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
2J4M
 
 | | Double dockerin from Piromyces equi Cel45A | | Descriptor: | ENDOGLUCANASE 45A | | Authors: | Nagy, T, Tunnicliffe, R.B, Higgins, L.D, Walters, C, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2006-09-01 | | Release date: | 2007-09-25 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Characterization of a Double Dockerin from the Cellulosome of the Anaerobic Fungus Piromyces Equi.
J.Mol.Biol., 373, 2007
|
|
2J4N
 
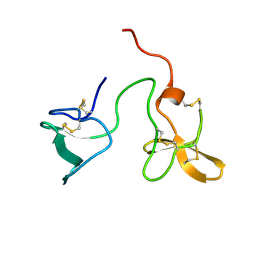 | | Double dockerin from Piromyces equi Cel45A | | Descriptor: | ENDOGLUCANASE 45A | | Authors: | Nagy, T, Tunnicliffe, R.B, Higgins, L.D, Walters, C, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2006-09-01 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Characterization of a Double Dockerin from the Cellulosome of the Anaerobic Fungus Piromyces Equi.
J.Mol.Biol., 373, 2007
|
|
1P82
 
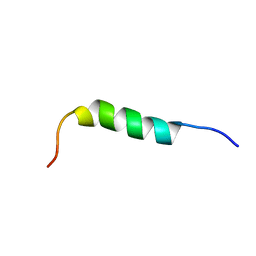 | | NMR STRUCTURE OF 1-25 FRAGMENT OF MYCOBACTERIUM TUBERCULOSIS CPN10 | | Descriptor: | 10 kDa chaperonin | | Authors: | Ciutti, A, Spiga, O, Giannozzi, E, Scarselli, M, Di Maro, D, Calamandrei, D, Niccolai, N, Bernini, A. | | Deposit date: | 2003-05-06 | | Release date: | 2003-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of 1-25 fragment of Cpn10 from Mycobacterium Tuberculosis
To be Published
|
|
1P83
 
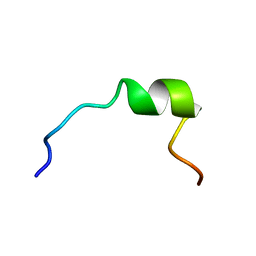 | | NMR STRUCTURE OF 1-25 FRAGMENT OF MYCOBACTERIUM TUBERCULOSIS CPN10 | | Descriptor: | 10 kDa chaperonin | | Authors: | Ciutti, A, Spiga, O, Giannozzi, E, Scarselli, M, Di Maro, D, Calamandrei, D, Niccolai, N, Bernini, A. | | Deposit date: | 2003-05-06 | | Release date: | 2003-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of 1-25 fragment of Cpn10 from Mycobacterium Tuberculosis
To be Published
|
|
