5ZR3
 
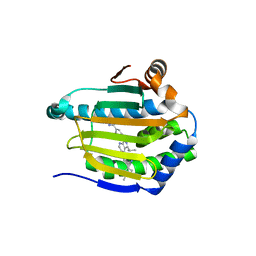 | | Crystal structure of Hsp90-alpha N-terminal domain in complex with 4-(3-isopropyl-4-(4-(1-methyl-1H-pyrazol-4-yl)-1H-imidazol-1-yl)-1H-pyrazolo[3,4-b]pyridin-1-yl)-3-methylbenzamide | | Descriptor: | 3-methyl-4-{4-[4-(1-methyl-1H-pyrazol-4-yl)-1H-imidazol-1-yl]-3-(propan-2-yl)-1H-pyrazolo[3,4-b]pyridin-1-yl}benzamide, Heat shock protein HSP 90-alpha | | Authors: | Uno, T, Chong, K.T, Suzuki, T. | | Deposit date: | 2018-04-23 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of 3-Ethyl-4-(3-isopropyl-4-(4-(1-methyl-1 H-pyrazol-4-yl)-1 H-imidazol-1-yl)-1 H-pyrazolo[3,4- b]pyridin-1-yl)benzamide (TAS-116) as a Potent, Selective, and Orally Available HSP90 Inhibitor.
J. Med. Chem., 62, 2019
|
|
5KEG
 
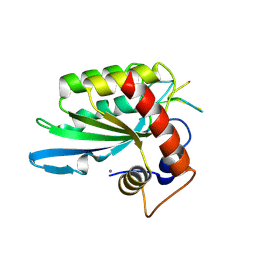 | | Crystal structure of APOBEC3A in complex with a single-stranded DNA | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*TP*TP*CP*TP*T)-3'), ... | | Authors: | Kouno, T, Hilbert, B.J, Silvas, T, Royer, W.E, Matsuo, H, Schiffer, C.A. | | Deposit date: | 2016-06-09 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity.
Nat Commun, 8, 2017
|
|
2RNG
 
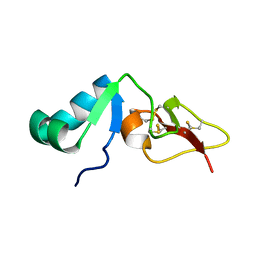 | | Solution structure of big defensin | | Descriptor: | Big defensin | | Authors: | Kouno, T, Fujitani, N, Osaki, T, Kawabata, S, Nishimura, S, Mizuguchi, M, Aizawa, T, Demura, M, Nitta, K, Kawano, K. | | Deposit date: | 2007-12-27 | | Release date: | 2008-10-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A novel beta-defensin structure: a potential strategy of big defensin for overcoming resistance by Gram-positive bacteria
Biochemistry, 47, 2008
|
|
2RQ2
 
 | | The solution structure of the N-terminal fragment of big defensin | | Descriptor: | Big defensin | | Authors: | Kouno, T, Mizuguchi, M, Aizawa, T, Shinoda, H, Demura, M, Kawabata, S, Kawano, K. | | Deposit date: | 2009-01-07 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel beta-defensin structure: big defensin changes its N-terminal structure to associate with the target membrane
Biochemistry, 48, 2009
|
|
8H0I
 
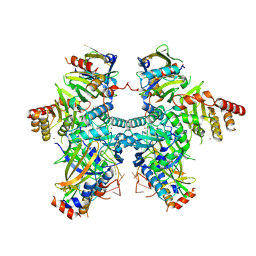 | | Cryo-EM structure of APOBEC3G-Vif complex | | Descriptor: | APOBEC3G, CHLORIDE ION, Core binding factor beta, ... | | Authors: | Kouno, T, Shibata, S, Hyun, J, Kim, T.G, Wolf, M. | | Deposit date: | 2022-09-29 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into RNA bridging between HIV-1 Vif and antiviral factor APOBEC3G.
Nat Commun, 14, 2023
|
|
8J62
 
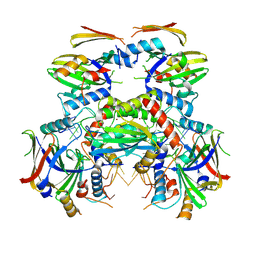 | | Cryo-EM structure of APOBEC3G-Vif complex | | Descriptor: | APOBEC3G, Core binding factor beta, RNA (5'-R(*CP*GP*GP*UP*UP*GP*AP*UP*UP*GP*UP*UP*UP*UP*AP*AP*CP*AP*A)-3'), ... | | Authors: | Kouno, T, Shibata, S, Hyun, J, Kim, T.G, Wolf, M. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into RNA bridging between HIV-1 Vif and antiviral factor APOBEC3G.
Nat Commun, 14, 2023
|
|
1V49
 
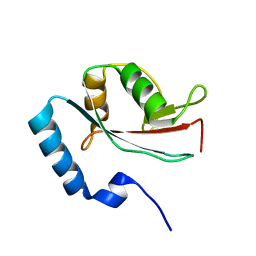 | | Solution structure of microtubule-associated protein light chain-3 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Kouno, T, Mizuguchi, M, Tanida, I, Ueno, T, Kominami, E, Kawano, K. | | Deposit date: | 2003-11-11 | | Release date: | 2004-12-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of microtubule-associated protein light chain 3 and identification of its functional subdomains.
J.Biol.Chem., 280, 2005
|
|
2JX8
 
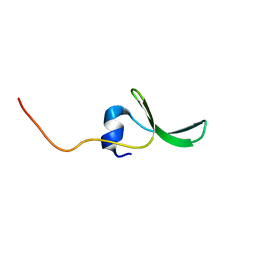 | | Solution structure of hPCIF1 WW domain | | Descriptor: | Phosphorylated CTD-interacting factor 1 | | Authors: | Kouno, T, Iwamoto, Y, Hirose, Y, Aizawa, T, Demura, M, Kawano, K, Ohkuma, Y, Mizuguchi, M. | | Deposit date: | 2007-11-09 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C, and 15N resonance assignments of hPCIF1 WW domain
To be Published
|
|
1KLV
 
 | | Solution Structure and Backbone Dynamics of GABARAP, GABAA Receptor associated protein | | Descriptor: | GABA(A) Receptor associated protein | | Authors: | Kouno, T, Miura, K, Tada, M, Kanematsu, T, Tate, S, Shirakawa, M, Hirata, M, Kawano, K. | | Deposit date: | 2001-12-13 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and '5N resonance assignments of GABARAP, GABAA receptor associated protein.
J.Biomol.Nmr, 22, 2002
|
|
1KM7
 
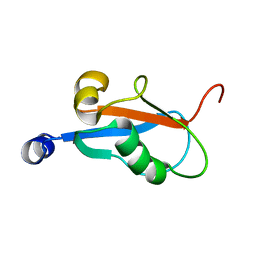 | | Solution Structure and Backbone Dynamics of GABARAP, GABAA Receptor Associated Protein | | Descriptor: | GABA(A) Receptor Associated Protein | | Authors: | Kouno, T, Miura, K, Tada, M, Kanematsu, T, Tate, S, Shirakawa, M, Hirata, M, Kawano, K. | | Deposit date: | 2001-12-14 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and '5N resonance assignments of GABARAP, GABAA receptor associated protein.
J.Biomol.Nmr, 22, 2002
|
|
2E2F
 
 | |
2MZZ
 
 | | NMR structure of APOBEC3G NTD variant, sNTD | | Descriptor: | Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G variant, ZINC ION | | Authors: | Kouno, T, Luengas, E.M, Shigematu, M, Shandilya, S.M.D, Zhang, J, Chen, L, Hara, M, Schiffer, C.A, Harris, R.S, Matsuo, H. | | Deposit date: | 2015-02-28 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Vif-binding domain of the antiviral enzyme APOBEC3G.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1V50
 
 | | Solution structure of phosphorylated N-terminal fragment of S100C/A11 protein | | Descriptor: | Calgizzarin | | Authors: | Kouno, T, Mizuguchi, M, Sakaguchi, M, Makino, E, Huh, N, Kawano, K. | | Deposit date: | 2003-11-20 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Study on structure-activity relationship between the N-terminal region of S100C protein and its function
Peptide Science, 40, 2003
|
|
1V4Z
 
 | | Solution structure of the N-terminal fragment of S100C/A11 protein | | Descriptor: | Calgizzarin | | Authors: | Kouno, T, Mizuguchi, M, Sakaguchi, M, Makino, E, Huh, N, Kawano, K. | | Deposit date: | 2003-11-20 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Study on structure-activity relationship between the N-terminal region of S100C protein and its function
Peptide Science, 40, 2003
|
|
2E45
 
 | |
2E8J
 
 | |
3A5P
 
 | | Crystal structure of hemagglutinin | | Descriptor: | Haemagglutinin I | | Authors: | Watanabe, N, Sakai, N, Nakamura, T, Nabeshima, Y, Kouno, T, Mizuguchi, M, Kawano, K. | | Deposit date: | 2009-08-10 | | Release date: | 2010-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Structure of Physarum polycephalum hemagglutinin I suggests a minimal carbohydrate recognition domain of legume lectin fold
J.Mol.Biol., 405, 2011
|
|
2KOZ
 
 | |
2KP0
 
 | |
3WQ9
 
 | | Crystal structure of Hsp90-alpha N-terminal domain in complex with 2-(4-Hydroxy-cyclohexylamino)-4-[5-(4-phenyl-imidazol-1-yl)-isoquinolin-1-yl]-benzamide | | Descriptor: | 2-[(trans-4-hydroxycyclohexyl)amino]-4-[5-(4-phenyl-1H-imidazol-1-yl)isoquinolin-1-yl]benzamide, Heat shock protein HSP 90-alpha | | Authors: | Chong, K.T, Yamashita, S, Oshiumi, H, Uno, T, Kitade, M. | | Deposit date: | 2014-01-23 | | Release date: | 2015-02-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of highly selective Hsp90 / inhibitors by structure and thermodynamics guided design
To be Published
|
|
3RI9
 
 | | Xylanase C from Aspergillus kawachii F131W mutant | | Descriptor: | Endo-1,4-beta-xylanase 3 | | Authors: | Fushinobu, S, Uno, T, Kitaoka, M, Hayashi, K, Matsuzawa, H, Wakagi, T. | | Deposit date: | 2011-04-13 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational analysis of fungal family 11 xylanases on pH optimum determination
J.APPL.GLYOSCI., 58, 2011
|
|
3RI8
 
 | | Xylanase C from Aspergillus kawachii D37N mutant | | Descriptor: | Endo-1,4-beta-xylanase 3 | | Authors: | Fushinobu, S, Uno, T, Kitaoka, M, Hayashi, K, Matsuzawa, H, Wakagi, T. | | Deposit date: | 2011-04-13 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational analysis of fungal family 11 xylanases on pH optimum determination
J.APPL.GLYOSCI., 58, 2011
|
|
7XJB
 
 | | Rat-COMT, opicapone,SAM and Mg bond | | Descriptor: | CHLORIDE ION, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Takebe, K, Iijima, H, Suzuki, M, Kuwada-Kusunose, T. | | Deposit date: | 2022-04-15 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Computational Analyses of the Unique Interactions of Opicapone in the Binding Pocket of Catechol O -Methyltransferase: A Crystallographic Study and Fragment Molecular Orbital Analyses.
J.Chem.Inf.Model., 63, 2023
|
|
4U4P
 
 | | Crystal structure of the human condensin SMC hinge domain heterodimer with short coiled coils | | Descriptor: | Structural maintenance of chromosomes protein 2, Structural maintenance of chromosomes protein 4 | | Authors: | Uchiyama, S, Kawahara, K, Hosokawa, Y, Fukakusa, S, Oki, H, Nakamura, S, Noda, M, Takino, R, Miyahara, Y, Maruno, T, Kobayashi, Y, Ohkubo, T, Fukui, K. | | Deposit date: | 2014-07-24 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for dimer information and DNA recognition of human SMC proteins
to be published
|
|
6LOS
 
 | | Crystal structure of mouse PEDF in complex with heterotrimeric collagen model peptide. | | Descriptor: | Collagen model peptide, type I, alpha 1, ... | | Authors: | Kawahara, K, Maruno, T, Oki, H, Yoshida, T, Ohkubo, T, Koide, T, Kobayashi, Y. | | Deposit date: | 2020-01-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.476 Å) | | Cite: | Spatiotemporal regulation of PEDF signaling by type I collagen remodeling.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
