5MWV
 
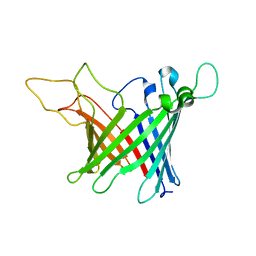 | | Solid-state NMR Structure of outer membrane protein G in lipid bilayers | | Descriptor: | Outer membrane protein G | | Authors: | Retel, J.S, Nieuwkoop, A.J, Hiller, M, Higman, V.A, Barbet-Massin, E, Stanek, J, Andreas, L.B, Franks, W.T, van Rossum, B.-J, Vinothkumar, K.R, Handel, L, de Palma, G.G, Bardiaux, B, Pintacuda, G, Emsley, L, Kuelbrandt, W, Oschkinat, H. | | Deposit date: | 2017-01-20 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure of outer membrane protein G in lipid bilayers.
Nat Commun, 8, 2017
|
|
6EKA
 
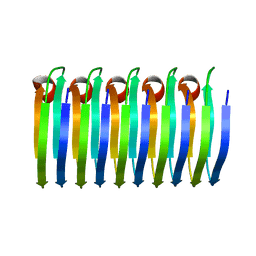 | | Solid-state MAS NMR structure of the HELLF prion amyloid fibrils | | Descriptor: | Podospora anserina S mat+ genomic DNA chromosome 3, supercontig 2 | | Authors: | Martinez, D, Daskalov, A, Andreas, L, Bardiaux, B, Coustou, V, Stanek, J, Berbon, M, Noubhani, M, Kauffmann, B, Wall, J.S, Pintacuda, G, Saupe, S.J, Habenstein, B, Loquet, A. | | Deposit date: | 2017-09-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structural and molecular basis of cross-seeding barriers in amyloids
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5JXV
 
 | | Solid-state MAS NMR structure of immunoglobulin beta 1 binding domain of protein G (GB1) | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Andreas, L.B, Jaudzems, K, Stanek, J, Lalli, D, Bertarello, A, Le Marchand, T, Cala-De Paepe, D, Kotelovica, S, Akopjana, I, Knott, B, Wegner, S, Engelke, F, Lesage, A, Emsley, L, Tars, K, Herrmann, T, Pintacuda, G. | | Deposit date: | 2016-05-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure of fully protonated proteins by proton-detected magic-angle spinning NMR.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JZR
 
 | | Solid-state MAS NMR structure of Acinetobacter phage 205 (AP205) coat protein in assembled capsid particles | | Descriptor: | Coat protein | | Authors: | Jaudzems, K, Andreas, L.B, Stanek, J, Lalli, D, Bertarello, A, Le Marchand, T, Cala-De Paepe, D, Kotelovica, S, Akopjana, I, Knott, B, Wegner, S, Engelke, F, Lesage, A, Emsley, L, Tars, K, Herrmann, T, Pintacuda, G. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure of fully protonated proteins by proton-detected magic-angle spinning NMR.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2NAR
 
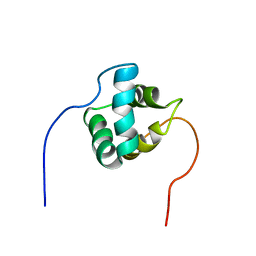 | | Solution structure of AVR3a_60-147 from Phytophthora infestans | | Descriptor: | Effector protein Avr3a | | Authors: | Matena, A, Bayer, P, Zhukov, I, Stanek, J, Kozminski, W, van West, P, Wawra, S. | | Deposit date: | 2016-01-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The RxLR Motif of the Host Targeting Effector AVR3a ofPhytophthora infestansIs Cleaved before Secretion.
Plant Cell, 29, 2017
|
|
2M3W
 
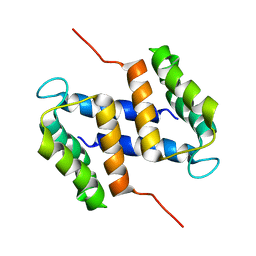 | |
6QAM
 
 | |
5FS4
 
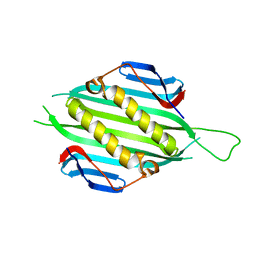 | | Bacteriophage AP205 coat protein | | Descriptor: | AP205 BACTERIOPHAGE COAT PROTEIN | | Authors: | Shishovs, M, Tars, K. | | Deposit date: | 2015-12-29 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure of Ap205 Coat Protein Reveals Circular Permutation in Ssrna Bacteriophages.
J.Mol.Biol., 428, 2016
|
|
5LQP
 
 | |
6QWR
 
 | |
