7ACG
 
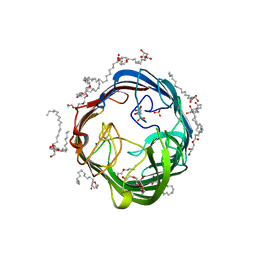 | | In meso structure of the alginate exporter, AlgE, from Pseudomonas aeruginosa in 9.8 monoacylglycerol | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-(2-METHOXYETHOXY)ETHANOL, Alginate biosynthesis protein AlgE, ... | | Authors: | Smithers, L, van Dalsen, L, Boland, C, Caffrey, M. | | Deposit date: | 2020-09-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 9.8 MAG. A new host lipid for in meso (lipid cubic phase) crystallization of integral membrane proteins
Cryst.Growth Des., 2020
|
|
7ACI
 
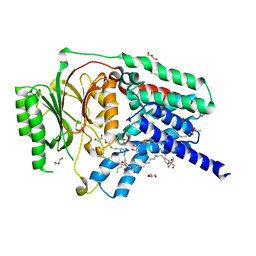 | | In meso structure of apolipoprotein N-acyltransferase, Lnt, from Escherichia coli in 9.8 monoacylglycerol | | Descriptor: | Apolipoprotein N-acyltransferase, GLYCEROL, [(2~{S})-2,3-bis(oxidanyl)propyl] heptadec-9-enoate | | Authors: | Smithers, L, van Dalsen, L, Boland, C, Caffrey, M. | | Deposit date: | 2020-09-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 9.8 MAG. A new host lipid for in meso (lipid cubic phase) crystallization of integral membrane proteins
Cryst.Growth Des., 2020
|
|
8RQP
 
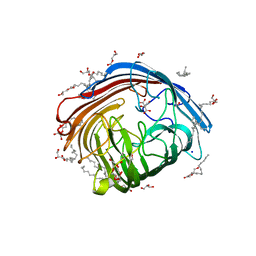 | | In meso structure of the alginate exporter, AlgE, from Pseudomonas aeruginosa in 7.10 monoacylglycerol | | Descriptor: | 7.10 monoacylglycerol (R-form), 7.10 monoacylglycerol (S-form), Alginate production protein AlgE, ... | | Authors: | Smithers, L, Boland, C, Krawinski, P, Caffrey, M. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 7.10 MAG. A Novel Host Monoacylglyceride for In Meso (Lipid Cubic Phase) Crystallization of Membrane Proteins.
Cryst.Growth Des., 24, 2024
|
|
8RQR
 
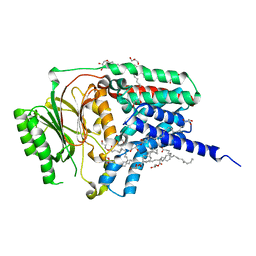 | | In meso structure of apolipoprotein N-acyltransferase, Lnt, from Escherichia coli in 7.10 monoacylglycerol | | Descriptor: | 7.10 monoacylglycerol (S-form), Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Smithers, L, Boland, C, Krawinski, P, Caffrey, M. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | 7.10 MAG. A Novel Host Monoacylglyceride for In Meso (Lipid Cubic Phase) Crystallization of Membrane Proteins.
Cryst.Growth Des., 24, 2024
|
|
8RQQ
 
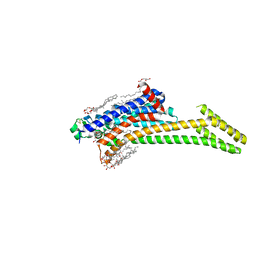 | | In meso structure of the adenosine A2a G protein-coupled receptor, A2aR, in 7.10 monoacylglycerol | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, 7.10 monoacylglycerol (R-form), 7.10 monoacylglycerol (S-form), ... | | Authors: | Smithers, L, Krawinski, P, Caffrey, M. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | 7.10 MAG. A Novel Host Monoacylglyceride for In Meso (Lipid Cubic Phase) Crystallization of Membrane Proteins.
Cryst.Growth Des., 24, 2024
|
|
8B0K
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli (Apo form) | | Descriptor: | Apolipoprotein N-acyltransferase | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8AQ2
 
 | | In meso structure of the membrane integral lipoprotein N-acyltransferase Lnt from P. aeruginosa covalently linked with TITC | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8AQ3
 
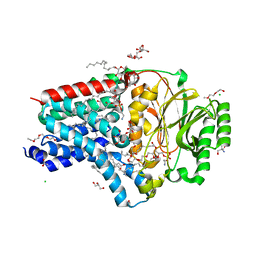 | | In surfo structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli in complex with PE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8AQ4
 
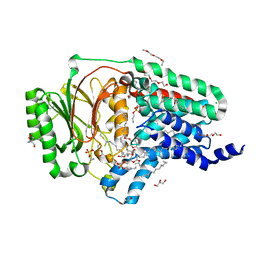 | | In surfo structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli in complex with TITC and lyso-PE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0N
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Lyso-PE | | Descriptor: | Apolipoprotein N-acyltransferase, [(2~{S})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-oxidanyl-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0P
 
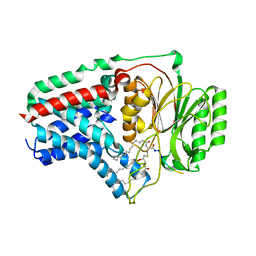 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Pam3 | | Descriptor: | Apolipoprotein N-acyltransferase, Pam3-SKKKK, [(2~{S})-3-[(2~{S})-3-azanyl-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0O
 
 | | Cryo-EM structure apolipoprotein N-acyltransferase Lnt from E.coli in complex with FP3 | | Descriptor: | Apolipoprotein N-acyltransferase, [(2~{R})-3-[(2~{R})-3-[[(2~{R})-1-[[(2~{R})-1-[[(2~{R})-6-[(2-aminophenyl)carbonylamino]-1-azanyl-1-oxidanylidene-hexan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0L
 
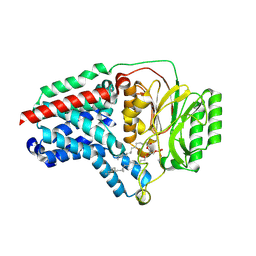 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE | | Descriptor: | Apolipoprotein N-acyltransferase, PHOSPHATIDYLETHANOLAMINE | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0M
 
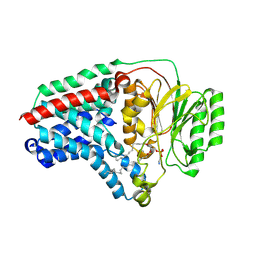 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE (C387S mutant) | | Descriptor: | Apolipoprotein N-acyltransferase, PHOSPHATIDYLETHANOLAMINE | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8UD0
 
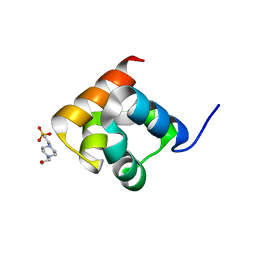 | |
8UCZ
 
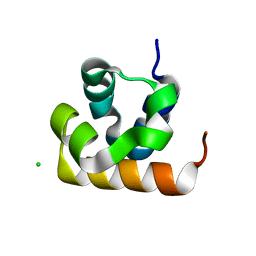 | |
8UCY
 
 | |
