6FY8
 
 | | The crystal structure of EncM bromide soak | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Saleem-Batcha, R, Teufel, R. | | Deposit date: | 2018-03-11 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Enzymatic control of dioxygen binding and functionalization of the flavin cofactor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FYA
 
 | |
6FOQ
 
 | |
6FOW
 
 | |
6FYC
 
 | |
6FYG
 
 | | The crystal structure of EncM V135T mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Saleem-Batcha, R, Teufel, R. | | Deposit date: | 2018-03-11 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Enzymatic control of dioxygen binding and functionalization of the flavin cofactor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FYB
 
 | | The crystal structure of EncM L144M mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Saleem-Batcha, R, Teufel, R. | | Deposit date: | 2018-03-11 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Enzymatic control of dioxygen binding and functionalization of the flavin cofactor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FYE
 
 | | The crystal structure of EncM H138T mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Saleem-Batcha, R, Teufel, R. | | Deposit date: | 2018-03-11 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic control of dioxygen binding and functionalization of the flavin cofactor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FYF
 
 | | The crystal structure of EncM V135M mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Saleem-Batcha, R, Teufel, R. | | Deposit date: | 2018-03-11 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic control of dioxygen binding and functionalization of the flavin cofactor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FP3
 
 | |
6FY9
 
 | |
6FYD
 
 | | The crystal structure of EncM T139V mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Saleem-Batcha, R, Teufel, R. | | Deposit date: | 2018-03-11 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Enzymatic control of dioxygen binding and functionalization of the flavin cofactor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8OVH
 
 | |
6TEF
 
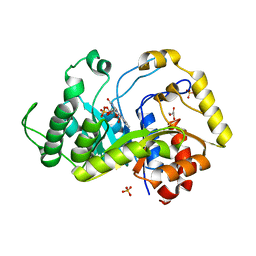 | | Crystal structure of monooxygenase RutA complexed with dioxygen under 0.5 MPa / 5 bars of oxygen pressure. | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Saleem-Batcha, R, Matthews, A, Teufel, R. | | Deposit date: | 2019-11-11 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Aminoperoxide adducts expand the catalytic repertoire of flavin monooxygenases.
Nat.Chem.Biol., 16, 2020
|
|
8COD
 
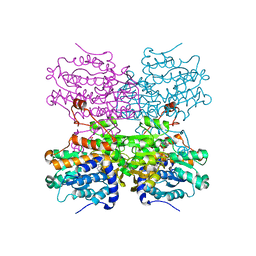 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Mus musculus in complex with inosine | | Descriptor: | Adenosylhomocysteinase, INOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Saleem-Batcha, R, Popadic, D, Koeppl, L.H, Andexer, J.N. | | Deposit date: | 2023-02-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure, function and substrate preferences of archaeal S-adenosyl-L-homocysteine hydrolases.
Commun Biol, 7, 2024
|
|
4B9H
 
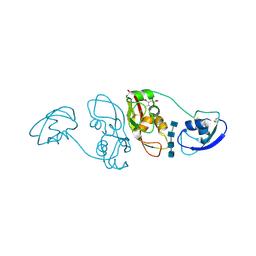 | | Cladosporium fulvum LysM effector Ecp6 in complex with a beta-1,4- linked N-acetyl-D-glucosamine tetramer: I3C heavy atom derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Saleem-Batcha, R, Sanchez-Vallet, A, Hansen, G, Thomma, B.P.H.J, Mesters, J.R. | | Deposit date: | 2012-09-04 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fungal Effector Ecp6 Outcompetes Host Immune Receptor for Chitin Binding Through Intrachain Lysm Dimerization
Elife, 2, 2013
|
|
7OUJ
 
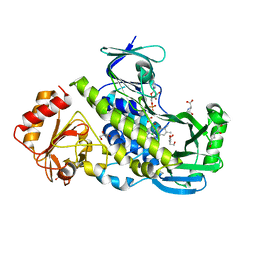 | | Crystal structure of the flavoprotein monooxygenase RubL from rubromycin biosynthesis | | Descriptor: | (2S)-hexane-1,2,6-triol, 4-HYDROXYPROLINE, CHLORIDE ION, ... | | Authors: | Saleem-Batcha, R, Toplak, M, Teufel, R. | | Deposit date: | 2021-06-11 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Catalytic Control of Spiroketal Formation in Rubromycin Polyketide Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7OUC
 
 | |
7OUD
 
 | |
4B8V
 
 | | Cladosporium fulvum LysM effector Ecp6 in complex with a beta-1,4- linked N-acetyl-D-glucosamine tetramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Saleem-Batcha, R, Sanchez-Vallet, A, Hansen, G, Thomma, B.P.H.J, Mesters, J.R. | | Deposit date: | 2012-08-30 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Fungal Effector Ecp6 Outcompetes Host Immune Receptor for Chitin Binding Through Intrachain Lysm Dimerization
Elife, 2, 2013
|
|
7R39
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Sulfolobus acidocaldarius in complex with adenosine | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Saleem-Batcha, R, Popadic, D, Andexer, J.N. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure, function and substrate preferences of archaeal S-adenosyl-L-homocysteine hydrolases.
Commun Biol, 7, 2024
|
|
7R3A
 
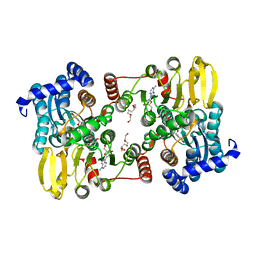 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Methanococcus maripaludis in complex with inosine | | Descriptor: | INOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-inosyl-L-homocysteine hydrolase, ... | | Authors: | Saleem-Batcha, R, Popadic, D, Andexer, J.N. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure, function and substrate preferences of archaeal S-adenosyl-L-homocysteine hydrolases.
Commun Biol, 7, 2024
|
|
7R37
 
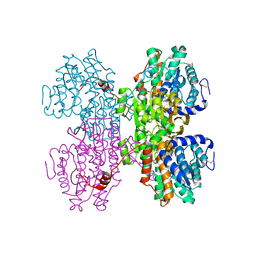 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Pyrococcus furiosus in complex with inosine | | Descriptor: | Adenosylhomocysteinase, INOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Saleem-Batcha, R, Popadic, D, Andexer, J.N. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure, function and substrate preferences of archaeal S-adenosyl-L-homocysteine hydrolases.
Commun Biol, 7, 2024
|
|
7R38
 
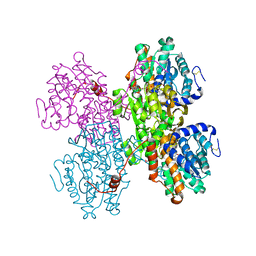 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Pyrococcus furiosus in complex with S-inosyl-L-homocysteine | | Descriptor: | (2S)-2-AMINO-4-({[(2S,3S,4R,5R)-3,4-DIHYDROXY-5-(6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)TETRAHYDROFURAN-2-YL]METHYL}THIO)BUTANOIC ACID, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Saleem-Batcha, R, Popadic, D, Andexer, J.N. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure, function and substrate preferences of archaeal S-adenosyl-L-homocysteine hydrolases.
Commun Biol, 7, 2024
|
|
8QNO
 
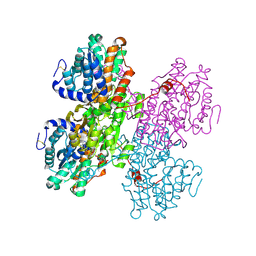 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase treated at 368 K from Pyrococcus furiosus in complex with inosine | | Descriptor: | Adenosylhomocysteinase, INOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Saleem-Batcha, R, Koeppl, L.H, Popadic, D, Andexer, J.N. | | Deposit date: | 2023-09-27 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Structure, function and substrate preferences of archaeal S-adenosyl-L-homocysteine hydrolases.
Commun Biol, 7, 2024
|
|
