1W3W
 
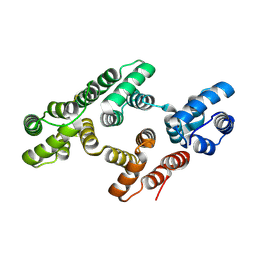 | | The 2.1 Angstroem resolution structure of annexin A8 | | Descriptor: | ANNEXIN A8, CALCIUM ION | | Authors: | Rety, S, Sopkova-De Oliveira Santos, J, Renouard, M, Lewit-Bentley, A. | | Deposit date: | 2004-07-20 | | Release date: | 2005-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Crystal Structure of Annexin A8 is Similar to that of Annexin A3
J.Mol.Biol., 345, 2005
|
|
1W45
 
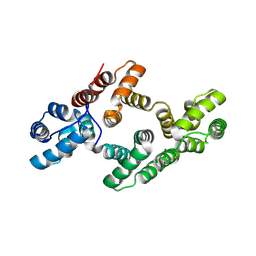 | | The 2.5 Angstroem structure of the K16A mutant of annexin A8, which has an intact N-terminus. | | Descriptor: | ANNEXIN A8 | | Authors: | Rety, S, Sopkova-de Oliveira Santos, J, Raguenes-Nicol, C, Dreyfuss, L, Blondeau, K, Hofbauerova, K, Renouard, M, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 2004-07-22 | | Release date: | 2005-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The Crystal Structure of Annexin A8 is Similar to that of Annexin A3
J.Mol.Biol., 345, 2005
|
|
1ANX
 
 | |
1BT6
 
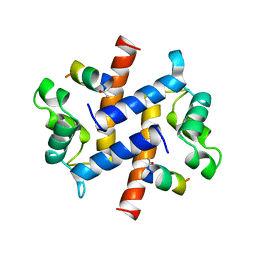 | | P11 (S100A10), LIGAND OF ANNEXIN II IN COMPLEX WITH ANNEXIN II N-TERMINUS | | Descriptor: | ANNEXIN II, S100A10 | | Authors: | Rety, S, Sopkova, J, Renouard, M, Osterloh, D, Gerke, V, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 1998-09-02 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of a complex of p11 with the annexin II N-terminal peptide.
Nat.Struct.Biol., 6, 1999
|
|
1A4P
 
 | | P11 (S100A10), LIGAND OF ANNEXIN II | | Descriptor: | S100A10 | | Authors: | Rety, S, Sopkova, J, Renouard, M, Osterloh, D, Gerke, V, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 1998-01-30 | | Release date: | 1998-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of a complex of p11 with the annexin II N-terminal peptide.
Nat.Struct.Biol., 6, 1999
|
|
4UW7
 
 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber without its intra-molecular chaperone domain | | Descriptor: | GLYCEROL, L-SHAPED TAIL FIBER PROTEIN | | Authors: | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-08-05 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
4UW8
 
 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber with its intra-molecular chaperone domain | | Descriptor: | CITRATE ANION, L-SHAPED TAIL FIBER PROTEIN | | Authors: | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
5AQ5
 
 | |
5TJT
 
 | |
5LXL
 
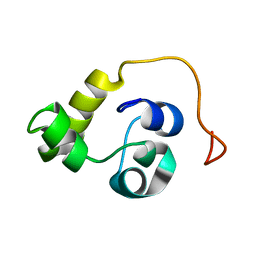 | | NMR structure of the N-terminal domain of the Bacteriophage T5 decoration protein pb10 | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
5LXK
 
 | | NMR structure of the C-terminal domain of the Bacteriophage T5 decoration protein pb10. | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
1QLS
 
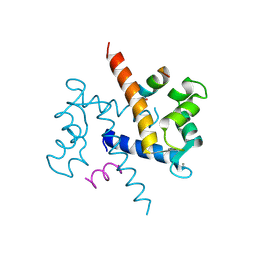 | | S100C (S100A11),OR CALGIZZARIN, IN COMPLEX WITH ANNEXIN I N-TERMINUS | | Descriptor: | ANNEXIN I, CALCIUM ION, S100C PROTEIN | | Authors: | Rety, S, Sopkova, J, Renouard, M, Osterloh, D, Gerke, V, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 1999-09-15 | | Release date: | 2000-02-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Ca2+ Dependent Association between S100C (S100A11) and its Target, the N-Terminal Part of Annexin I
Structure, 8, 2000
|
|
1OEJ
 
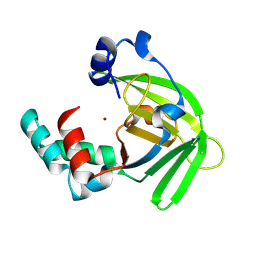 | | YodA from Escherichia coli crystallised with no added ions | | Descriptor: | HYPOTHETICAL PROTEIN YODA, NICKEL (II) ION | | Authors: | David, G, Blondeau, K, Renouard, M, Penel, S, Lewit-Bentley, A. | | Deposit date: | 2003-03-27 | | Release date: | 2003-08-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Yoda from Escherichia Coli is a Metal-Binding, Lipocalin-Like Protein
J.Biol.Chem., 278, 2003
|
|
1OEE
 
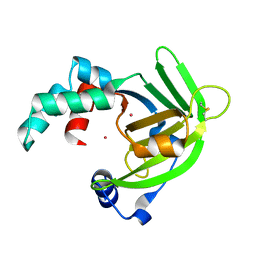 | | YodA from Escherichia coli crystallised with cadmium ions | | Descriptor: | CADMIUM ION, HYPOTHETICAL PROTEIN YODA | | Authors: | David, G, Blondeau, K, Renouard, M, Penel, S, Lewit-Bentley, A. | | Deposit date: | 2003-03-27 | | Release date: | 2003-08-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Yoda from Escherichia Coli is a Metal-Binding, Lipocalin-Like Protein
J.Biol.Chem., 278, 2003
|
|
1OEK
 
 | | YodA from Escherichia coli crystallised with zinc ions | | Descriptor: | METAL-BINDING PROTEIN ZINT, ZINC ION | | Authors: | David, G, Blondeau, K, Renouard, M, Penel, S, Lewit-Bentley, A. | | Deposit date: | 2003-03-28 | | Release date: | 2003-08-15 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Yoda from Escherichia Coli is a Metal-Binding, Lipocalin-Like Protein
J.Biol.Chem., 278, 2003
|
|
