6XHH
 
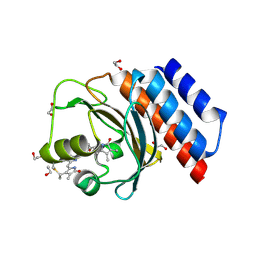 | | Far-red absorbing dark state of JSC1_58120g3 with bound 18-1, 18-2 dihydrobiliverdin IXa (DHBV), the native chromophore precursor | | Descriptor: | 1,2-ETHANEDIOL, JSC1_58120g3, mesobiliverdin IX(alpha) | | Authors: | Moreno, M.V, Rockwell, N.C, Fisher, A.J, Lagarias, J.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A far-red cyanobacteriochrome lineage specific for verdins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XHG
 
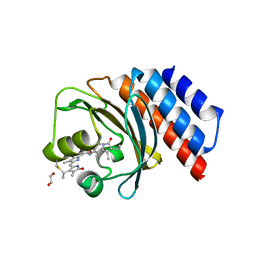 | | Far-red absorbing dark state of JSC1_58120g3 with bound biliverdin IXa (BV) | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(~{Z})-[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, JSC1_58120g3 | | Authors: | Moreno, M.V, Rockwell, N.C, Fisher, A.J, Lagarias, J.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A far-red cyanobacteriochrome lineage specific for verdins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UVB
 
 | |
6UV8
 
 | |
6BTF
 
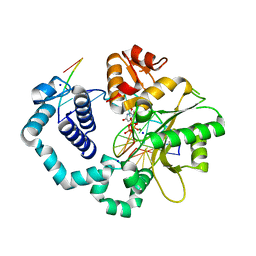 | | DNA Polymerase Beta I260Q Ternary Complex | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA Downstream Strand, DNA Primer Strand, ... | | Authors: | Eckenroth, B.E, Doublie, S. | | Deposit date: | 2017-12-06 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | I260Q DNA polymerase beta highlights precatalytic conformational rearrangements critical for fidelity.
Nucleic Acids Res., 46, 2018
|
|
6BTE
 
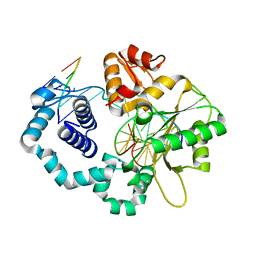 | | DNA Polymerase Beta I260Q Binary Complex | | Descriptor: | CHLORIDE ION, DNA Downstream Strand, DNA Primer Strand, ... | | Authors: | Eckenroth, B.E, Doublie, S. | | Deposit date: | 2017-12-06 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | I260Q DNA polymerase beta highlights precatalytic conformational rearrangements critical for fidelity.
Nucleic Acids Res., 46, 2018
|
|
