4UX9
 
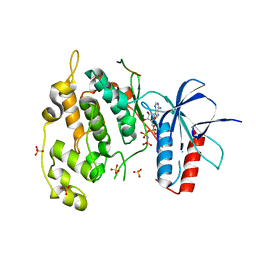 | | Crystal structure of JNK1 bound to a MKK7 docking motif | | Descriptor: | DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 7, MITOGEN-ACTIVATED PROTEIN KINASE 8, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Kragelj, J, Palencia, A, Nanao, M.H, Maurin, D, Bouvignies, G, Blackledge, M, Ringkjobing-Jensen, M. | | Deposit date: | 2014-08-20 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and Dynamics of the Mkk7-Jnk Signaling Complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6H5S
 
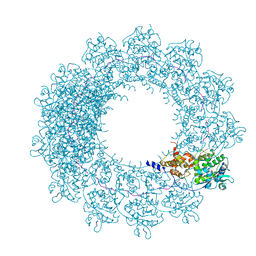 | | Cryo-EM map of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to viral genomic 5-prime RNA hexamers. | | Descriptor: | Nucleocapsid, RNA (5'-R(*AP*CP*CP*AP*GP*A)-3') | | Authors: | Desfosses, A, Milles, S, Ringkjobing Jensen, M, Guseva, S, Colletier, J.P, Maurin, D, Schoehn, G, Gutsche, I, Ruigrok, R, Blackledge, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2BYO
 
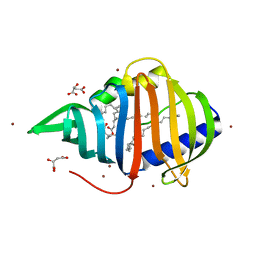 | | Crystal structure of Mycobacterium tuberculosis lipoprotein LppX (Rv2945c) | | Descriptor: | ACETATE ION, ALPHA-LINOLENIC ACID, D-MALATE, ... | | Authors: | Sulzenbacher, G, Canaan, S, Roig-Zamboni, V, Maurin, D, Gicquel, B, Bourne, Y. | | Deposit date: | 2005-08-03 | | Release date: | 2006-03-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Lppx is a Lipoprotein Required for the Translocation of Phthiocerol Dimycocerosates to the Surface of Mycobacterium Tuberculosis.
Embo J., 25, 2006
|
|
6H5Q
 
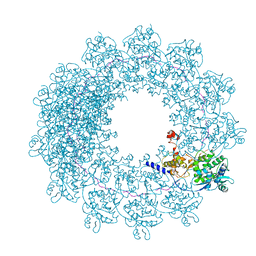 | | Cryo-EM structure of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to polyA RNA hexamers. | | Descriptor: | Nucleocapsid, RNA (5'-R(*AP*AP*AP*AP*AP*A)-3') | | Authors: | Desfosses, A, Milles, S, Ringkjobing Jensen, M, Guseva, S, Colletier, J, Maurin, D, Schoehn, G, Gutsche, I, Ruigrok, R, Blackledge, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1W9A
 
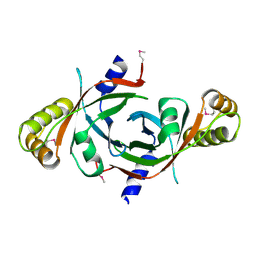 | | Crystal structure of Rv1155 from Mycobacterium tuberculosis | | Descriptor: | PUTATIVE PYRIDOXINE/PYRIDOXAMINE 5'-PHOSPHATE OXIDASE | | Authors: | Cannan, S, Sulzenbacher, G, Roig-Zamboni, V, Scappuccini, L, Frassinetti, F, Maurien, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2004-10-07 | | Release date: | 2005-01-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Conserved Hypothetical Protein Rv1155 from Mycobacterium Tuberculosis
FEBS Lett., 579, 2005
|
|
7PKU
 
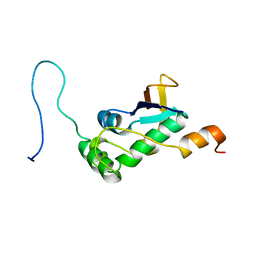 | | Structure of SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a | | Descriptor: | 3C-like proteinase, Nucleoprotein | | Authors: | Bessa, L.M, Guseva, S, Camacho-Zarco, A.R, Salvi, N, Blackledge, M. | | Deposit date: | 2021-08-26 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The intrinsically disordered SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a.
Sci Adv, 8, 2022
|
|
3ZDO
 
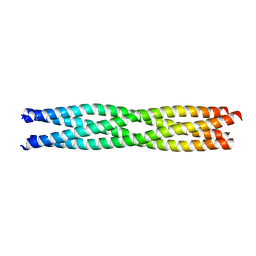 | | Tetramerization domain of Measles virus phosphoprotein | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PHOSPHOPROTEIN | | Authors: | Communie, G, Crepin, T, Jensen, M.R, Blackledge, M, Ruigrok, R.W.H. | | Deposit date: | 2012-11-29 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of the Tetramerization Domain of Measles Virus Phosphoprotein.
J.Virol., 87, 2013
|
|
7NYK
 
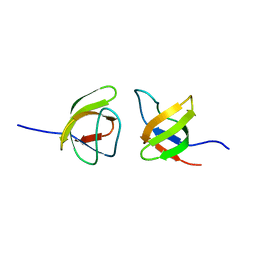 | |
7NYM
 
 | | Mutant V517A - SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | HEXAETHYLENE GLYCOL, PHOSPHATE ION, SH3 domain of JNK-interacting Protein 1 (JIP1), ... | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.614 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NZB
 
 | | Mutant V517L of the SH3 domain of JNK-interacting protein 1 (JIP1) | | Descriptor: | PHOSPHATE ION, SH3 domain of JNK-interacting protein 1 (JIP1), TETRAETHYLENE GLYCOL | | Authors: | Perez, L.M, Ielasi, F.S, Jensen, M.R, Palencia, A. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NYN
 
 | | Mutant Y526A of SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.537 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NYL
 
 | | Mutant H493A of SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | SH3 domain of JNK-interacting Protein 1 (JIP1), TETRAETHYLENE GLYCOL, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NZD
 
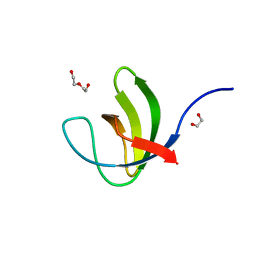 | | Fourth SH3 domain of POSH (Plenty of SH3 Domains protein) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase SH3RF1 | | Authors: | Palencia, A, Bessa, L.M, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NYO
 
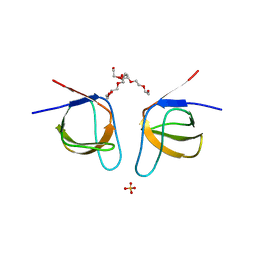 | | Mutant A541L of SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | 1,2-ETHANEDIOL, SH3 domain of JNK-interacting Protein 1 (JIP1), SULFATE ION, ... | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NZC
 
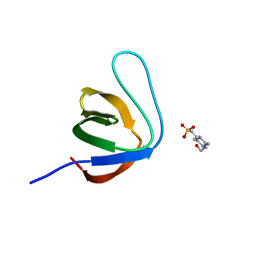 | |
