4ABT
 
 | | Crystal structure of Type IIF restriction endonuclease NgoMIV with cognate uncleaved DNA | | Descriptor: | 5'-D(*TP*GP*CP*GP*CP*CP*GP*GP*CP*GP*CP)-3', CALCIUM ION, TYPE-2 RESTRICTION ENZYME NGOMIV | | Authors: | Manakova, E.N, Grazulis, S, Zaremba, M, Tamulaitiene, G, Golovenko, D, Siksnys, V. | | Deposit date: | 2011-12-11 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure of Type Iif Restriction Endonuclease Ngomiv with Cognate Uncleaved DNA
To be Published
|
|
8QG0
 
 | | Archaeoglobus fulgidus AfAgo complex with AfAgo-N protein (fAfAgo) bound with 17 nt RNA guide and 17 nt DNA target | | Descriptor: | AfAgo-N protein, DNA target 17 nt, Piwi protein, ... | | Authors: | Manakova, E.N, Zaremba, M, Pocevicuite, R, Golovinas, E, Zagorskaite, E, Silanskas, A. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res., 52, 2024
|
|
8OLD
 
 | | Crystal structure of Archaeoglobus fulgidus AfAgo-N protein representing N-L1-L2 domains | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Archaeoglobus fulgidus AfAgo-N protein representing N-L1-L2 domains, CACODYLATE ION, ... | | Authors: | Manakova, E.N, Zaremba, M, Grazulis, S. | | Deposit date: | 2023-03-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res., 52, 2024
|
|
8OK9
 
 | |
8OLJ
 
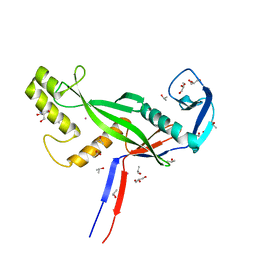 | | Crystal structure of Archaeoglobus fulgidus AfAgo-N protein representing N-L1-L2 domains | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, Archaeoglobus fulgidus AfAgo-N protein representing N-L1-L2 domains, ... | | Authors: | Manakova, E.N, Zaremba, M, Grazulis, S. | | Deposit date: | 2023-03-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res., 52, 2024
|
|
8PVV
 
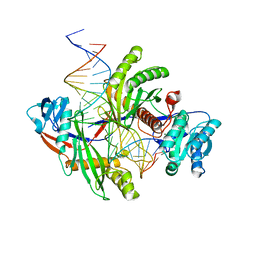 | | Archaeoglobus fulgidus AfAgo complex with AfAgo-N protein (fAfAgo) bound with 30 nt RNA guide and 51 nt DNA target | | Descriptor: | Archaeoglobus fulgidus AfAgo-N protein, DNA (51-MER), MAGNESIUM ION, ... | | Authors: | Manakova, E.N, Zaremba, M, Pocevicuite, R, Golovinas, E, Sasnauskas, G, Zagorskaite, E, Silanskas, A. | | Deposit date: | 2023-07-18 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res., 52, 2024
|
|
8RAR
 
 | | Crystal structure of chimeric human carbonic anhydrase IX with N-butyl-4-chloro-2-(cyclohexylsulfanyl)-5-sulfamoylbenzamide | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ZINC ION, ... | | Authors: | Manakova, E.N, Smirnov, A, Paketuryte, V, Grazulis, S. | | Deposit date: | 2023-12-01 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | From X-ray crystallographic structure to intrinsic thermodynamics of protein-ligand binding using carbonic anhydrase isozymes as a model system.
Iucrj, 11, 2024
|
|
8RBP
 
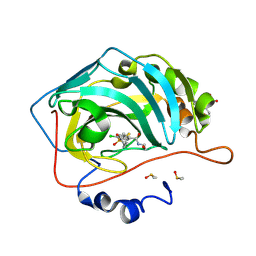 | | Crystal structure of chimeric human carbonic anhydrase IX with 4-chloro-2-(cyclohexylsulfanyl)-N-(2-hydroxyethyl)-5-sulfamoylbenzamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-chloranyl-2-cyclohexylsulfanyl-~{N}-(2-hydroxyethyl)-5-sulfamoyl-benzamide, Carbonic anhydrase 2, ... | | Authors: | Manakova, E.N, Smirnov, A, Paketuryte, V, Grazulis, S. | | Deposit date: | 2023-12-04 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | From X-ray crystallographic structure to intrinsic thermodynamics of protein-ligand binding using carbonic anhydrase isozymes as a model system.
Iucrj, 11, 2024
|
|
3V1Z
 
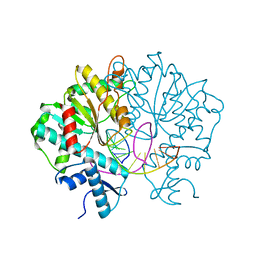 | | Crystal structure of Type IIF restriction endonuclease Bse634I with cognate DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*CP*GP*GP*CP*GP*CP*G)-3'), Endonuclease Bse634IR | | Authors: | Manakova, E.N, Grazulis, S, Golovenko, D, Tamulaitiene, G. | | Deposit date: | 2011-12-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural mechanisms of the degenerate sequence recognition by Bse634I restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
3V20
 
 | | Crystal structure of Type IIF restriction endonuclease Bse634I with cognate DNA | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Manakova, E.N, Grazulis, S, Golovenko, D, Tamulaitiene, G. | | Deposit date: | 2011-12-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural mechanisms of the degenerate sequence recognition by Bse634I restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
3V21
 
 | | Crystal structure of Type IIF restriction endonuclease Bse634I with cognate DNA | | Descriptor: | DNA (5'-D(*TP*TP*CP*GP*AP*CP*CP*GP*GP*TP*CP*GP*A)-3'), Endonuclease Bse634IR | | Authors: | Manakova, E.N, Grazulis, S, Golovenko, D, Tamulaitiene, G. | | Deposit date: | 2011-12-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural mechanisms of the degenerate sequence recognition by Bse634I restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
8RJ2
 
 | | Crystal structure of carbonic anhydrase II with N-butyl-4-chloro-2-(cyclohexylsulfanyl)-5-sulfamoylbenzamide | | Descriptor: | BICINE, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E.N, Grazulis, S. | | Deposit date: | 2023-12-19 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | From X-ray crystallographic structure to intrinsic thermodynamics of protein-ligand binding using carbonic anhydrase isozymes as a model system.
Iucrj, 11, 2024
|
|
