1OJH
 
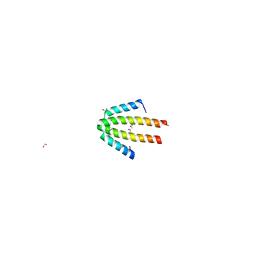 | | Crystal structure of NblA from PCC 7120 | | Descriptor: | 1,2-ETHANEDIOL, NBLA | | Authors: | Bienert, R, Baier, K, Lockau, W, Heinemann, U. | | Deposit date: | 2003-07-10 | | Release date: | 2004-07-15 | | Last modified: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Nbla from Anabaena Sp. Pcc 7120, a Small Protein Playing a Key Role in Phycobilisome Degradation.
J.Biol.Chem., 281, 2006
|
|
1T3M
 
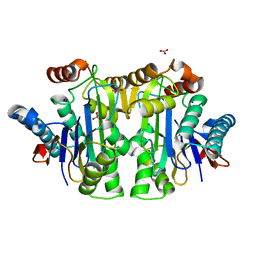 | | Structure of the isoaspartyl peptidase with L-asparaginase activity from E. coli | | Descriptor: | NITRATE ION, Putative L-asparaginase, SODIUM ION | | Authors: | Prahl, A, Pazgier, M, Hejazi, M, Lockau, W, Lubkowski, J. | | Deposit date: | 2004-04-27 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the isoaspartyl peptidase with L-asparaginase activity from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2GEZ
 
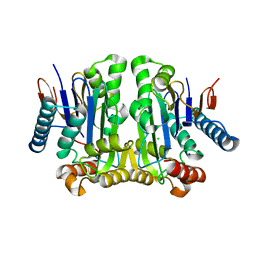 | | Crystal structure of potassium-independent plant asparaginase | | Descriptor: | CHLORIDE ION, L-asparaginase alpha subunit, L-asparaginase beta subunit, ... | | Authors: | Michalska, K, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-03-21 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of plant asparaginase.
J.Mol.Biol., 360, 2006
|
|
2ZAK
 
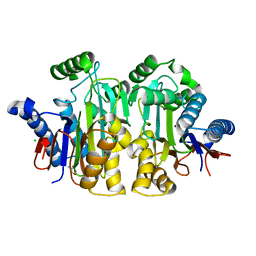 | | Orthorhombic crystal structure of precursor E. coli isoaspartyl peptidase/L-asparaginase (EcAIII) with active-site T179A mutation | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, L-asparaginase precursor, ... | | Authors: | Michalska, K, Hernandez-Santoyo, A, Jaskolski, M. | | Deposit date: | 2007-10-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal packing of plant-type L-asparaginase from Escherichia coli
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3C17
 
 | |
2ZAL
 
 | | Crystal structure of E. coli isoaspartyl aminopeptidase/L-asparaginase in complex with L-aspartate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ASPARTIC ACID, CALCIUM ION, ... | | Authors: | Michalska, K, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2007-10-07 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of isoaspartyl aminopeptidase in complex with L-aspartate
J.Biol.Chem., 280, 2005
|
|
