1DRR
 
 | | DNA/RNA HYBRID DUPLEX CONTAINING A PURINE-RICH DNA STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*AP*GP*AP*AP*GP*C)-3'), RNA (5'-R(*GP*CP*UP*UP*CP*UP*CP*UP*UP*C)-3') | | Authors: | Gyi, J.I, Lane, A.N, Conn, G.L, Brown, T. | | Deposit date: | 1997-10-21 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes.
Biochemistry, 37, 1998
|
|
1K0R
 
 | | Crystal Structure of Mycobacterium tuberculosis NusA | | Descriptor: | NusA, SULFATE ION | | Authors: | Gopal, B, Haire, L.F, Gamblin, S.J, Dodson, E.J, Lane, A.N, Papavinasasundaram, K.G, Colston, M.J, Dodson, G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-09-20 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the transcription elongation/anti-termination factor NusA from Mycobacterium tuberculosis at 1.7 A resolution.
J.Mol.Biol., 314, 2001
|
|
1L3G
 
 | | NMR Structure of the DNA-binding Domain of Cell Cycle Protein, Mbp1(2-124) from Saccharomyces cerevisiae | | Descriptor: | TRANSCRIPTION FACTOR Mbp1 | | Authors: | Nair, M, McIntosh, P.B, Frenkiel, T.A, Kelly, G, Taylor, I.A, Smerdon, S.J, Lane, A.N. | | Deposit date: | 2002-02-27 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the DNA-Binding Domain of the Cell Cycle Protein Mbp1 from Saccharomyces cerevisiae
Biochemistry, 42, 2003
|
|
1OO7
 
 | | DNA.RNA HYBRID DUPLEX CONTAINING A 5-PROPYNE DNA STRAND AND PURINE-RICH RNA STRAND, NMR, 4 STRUCTURES | | Descriptor: | 5'-D(*GP*(5PC)P*(PDU)P*(PDU)P*(5PC)P*(PDU)P*(5PC)P*(PDU)P*(PDU)P*C)-3', 5'-R(*GP*AP*AP*GP*AP*GP*AP*AP*GP*C)-3' | | Authors: | Gyi, J.I, Gao, D, Conn, G.L, Trent, J.O, Brown, T, Lane, A.N. | | Deposit date: | 2003-03-03 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a DNA*RNA duplex containing 5-propynyl U and C; comparison with 5-Me modifications
Nucleic Acids Res., 31, 2003
|
|
2MHY
 
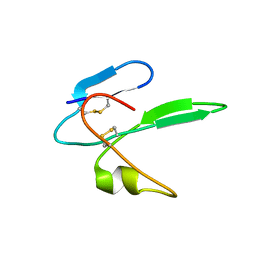 | | Structure determination of the salamander courtship pheromone Plethodontid Modulating Factor | | Descriptor: | Plethodontid modulating factor | | Authors: | Wilburn, D.B, Bowen, K.E, Doty, K.A, Arumugam, S, Lane, A.N, Feldhoff, P.W, Feldhoff, R.C. | | Deposit date: | 2013-12-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the evolution of a sexy protein: novel topology and restricted backbone flexibility in a hypervariable pheromone from the red-legged salamander, Plethodon shermani.
Plos One, 9, 2014
|
|
2KRR
 
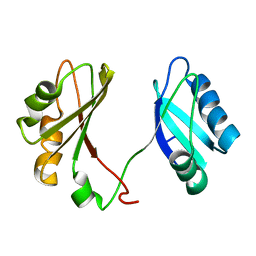 | | Solution structure of the RBD1,2 domains from human nucleolin | | Descriptor: | Nucleolin | | Authors: | Arumugam, N, Miller, C, Maliekal, J, Bates, P.J, Trent, J.O, Lane, A.N. | | Deposit date: | 2009-12-22 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RBD1,2 domains from human nucleolin.
J.Biomol.Nmr, 47, 2010
|
|
1RRD
 
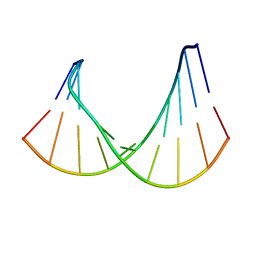 | | DNA/RNA HYBRID DUPLEX CONTAINING A PURINE-RICH RNA STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*CP*TP*CP*TP*TP*C)-3'), RNA (5'-R(*GP*AP*AP*GP*AP*GP*AP*AP*GP*C)-3') | | Authors: | Gyi, J.I, Lane, A.N, Conn, G.L, Brown, T. | | Deposit date: | 1997-10-21 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes.
Biochemistry, 37, 1998
|
|
1RRR
 
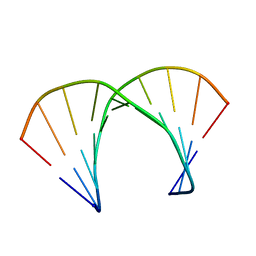 | | RNA DUPLEX CONTAINING A PURINE-RICH STRAND, NMR, 10 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*AP*AP*GP*AP*GP*AP*AP*GP*C)-3'), RNA (5'-R(*GP*CP*UP*UP*CP*UP*CP*UP*UP*C)-3') | | Authors: | Gyi, J.I, Lane, A.N, Conn, G.L, Brown, T. | | Deposit date: | 1997-10-21 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes.
Biochemistry, 37, 1998
|
|
2RN4
 
 | |
1SAA
 
 | | ATF-2 RECOGNITION SITE, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*TP*GP*AP*CP*GP*TP*CP*AP*CP*AP*TP*G)-3') | | Authors: | Conte, M.R, Lane, A.N, Bloomberg, G. | | Deposit date: | 1997-08-19 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ATF-2 recognition site and its interaction with the ATF-2 peptide.
Nucleic Acids Res., 25, 1997
|
|
1AXP
 
 | | DNA DUPLEX CONTAINING A PURINE-RICH STRAND, NMR, 6 STRUCTURES | | Descriptor: | DNA (D(GAAGAGAAGC)(DOT)D(GCTTCTCTTC)) | | Authors: | Gyi, J.I, Lane, A.N, Conn, G.L, Brown, T. | | Deposit date: | 1997-10-17 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes.
Biochemistry, 37, 1998
|
|
1AL5
 
 | | A-TRACT RNA DODECAMER, NMR, 12 STRUCTURES | | Descriptor: | RNA (5'-R(*CP*GP*CP*AP*AP*AP*UP*UP*UP*GP*CP*G)-3') | | Authors: | Conte, M.R, Conn, G.L, Brown, T, Lane, A.N. | | Deposit date: | 1997-06-11 | | Release date: | 1997-12-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational properties and thermodynamics of the RNA duplex r(CGCAAAUUUGCG)2: comparison with the DNA analogue d(CGCAAATTTGCG)2.
Nucleic Acids Res., 25, 1997
|
|
1BCE
 
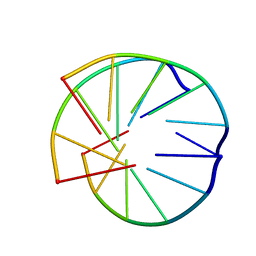 | | INTRAMOLECULAR TRIPLEX, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*AP*GP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*CP*TP*T)-3') | | Authors: | Asensio, J.L, Brown, T, Lane, A.N. | | Deposit date: | 1998-04-29 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Comparison of the solution structures of intramolecular DNA triple helices containing adjacent and non-adjacent CG.C+ triplets.
Nucleic Acids Res., 26, 1998
|
|
1BWG
 
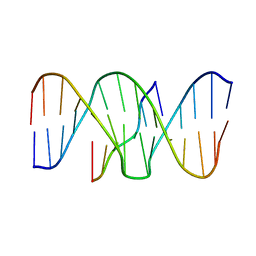 | | DNA TRIPLEX WITH 5' AND 3' JUNCTIONS, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*CP*T)-3'), DNA (5'-D(*GP*AP*CP*TP*GP*AP*GP*AP*GP*AP*CP*GP*TP*A)-3'), DNA (5'-D(*TP*AP*CP*GP*TP*CP*TP*CP*TP*CP*AP*GP*TP*C)-3') | | Authors: | Asensio, J.L, Brown, T, Lane, A.N. | | Deposit date: | 1998-09-22 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of a parallel DNA triple helix with 5' and 3' triplex-duplex junctions.
Structure Fold.Des., 7, 1999
|
|
1BCB
 
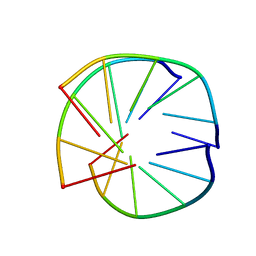 | | INTRAMOLECULAR TRIPLEX, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*GP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*CP*T)-3') | | Authors: | Asensio, J.L, Brown, T, Lane, A.N. | | Deposit date: | 1998-04-29 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Comparison of the solution structures of intramolecular DNA triple helices containing adjacent and non-adjacent CG.C+ triplets.
Nucleic Acids Res., 26, 1998
|
|
1E52
 
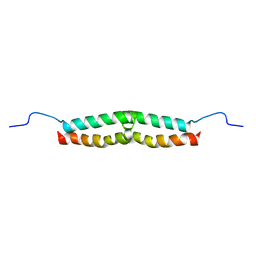 | | Solution structure of Escherichia coli UvrB C-terminal domain | | Descriptor: | EXCINUCLEASE ABC SUBUNIT | | Authors: | Alexandrovich, A.A, Kelly, G.G, Frenkiel, T.A, Moolenaar, G.F, Goosen, N.N, Sanderson, M.R, Lane, A.N. | | Deposit date: | 2000-07-14 | | Release date: | 2001-07-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Hydrodynamics and Thermodynamics of the Uvrb C-Terminal Domain.
J.Biomol.Struct.Dyn., 19, 2001
|
|
5MVL
 
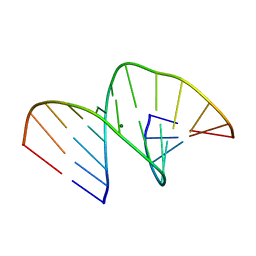 | | Crystal structure of an A-DNA dodecamer containing 5-bromouracil | | Descriptor: | Brominated DNA dodecamer, MAGNESIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-16 | | Release date: | 2017-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.405 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6QJR
 
 | | Crystal structure of a DNA dodecamer containing a tetramethylpyrrolinoxyl (nitroxide) spin label | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*(5NO)P*GP*CP*G)-3') | | Authors: | Hardwick, J.S, Haugland, M.M, Ptchelkine, D, Brown, T, Anderson, E.E. | | Deposit date: | 2019-01-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2'-Alkynyl spin-labelling is a minimally perturbing tool for DNA structural analysis.
Nucleic Acids Res., 48, 2020
|
|
6QJS
 
 | | Crystal structure of a DNA dodecamer containing a tetramethylpiperidinoxyl (nitroxide) spin label | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(P*GP*CP*AP*AP*AP*TP*TP*(S6M)P*GP*CP*G)-3') | | Authors: | Hardwick, J.S, Haugland, M.M, Ptchelkine, D, Brown, T, Anderson, E.E. | | Deposit date: | 2019-01-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 2'-Alkynyl spin-labelling is a minimally perturbing tool for DNA structural analysis.
Nucleic Acids Res., 48, 2020
|
|
1AO9
 
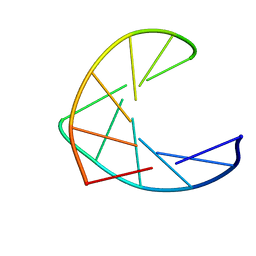 | |
1AT4
 
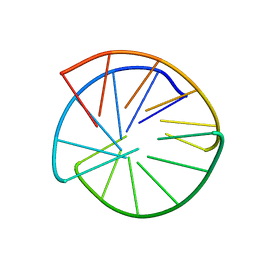 | |
5MVT
 
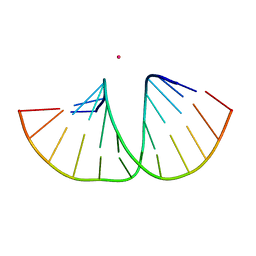 | | Crystal structure of an A-DNA dodecamer featuring an alternating pyrimidine-purine sequence | | Descriptor: | COBALT (III) ION, DNA | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVQ
 
 | | Crystal structure of an unmodified, self-complementary dodecamer. | | Descriptor: | DNA, MAGNESIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVU
 
 | |
5MVP
 
 | | Crystal structure of an A-DNA dodecamer containing the GGGCCC motif | | Descriptor: | DNA, POTASSIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
