1AIV
 
 | | APO OVOTRANSFERRIN | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, OVOTRANSFERRIN | | Authors: | Kurokawa, H, Dewan, J.C, Mikami, B, Sacchettini, J.C, Hirose, M. | | Deposit date: | 1997-04-28 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of hen apo-ovotransferrin. Both lobes adopt an open conformation upon loss of iron
J.Biol.Chem., 274, 1999
|
|
3A5T
 
 | | Crystal structure of MafG-DNA complex | | Descriptor: | 5'-D(*CP*TP*GP*AP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*TP*CP*AP*G)-3', MAGNESIUM ION, ... | | Authors: | Kurokawa, H, Motohashi, H, Sueno, S, Kimura, M, Takagawa, H, Kanno, Y, Yamamoto, M, Tanaka, T. | | Deposit date: | 2009-08-11 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Alternative DNA Recognition by Maf Transcription Factors
Mol.Cell.Biol., 29, 2009
|
|
7CPM
 
 | | CRYSTAL STRUCTURE OF DODECAPRENYL DIPHOSPHATE SYNTHASE FROM THERMOBIFIDA FUSCA | | Descriptor: | Trans,polycis-polyprenyl diphosphate synthase ((2Z,6E)-farnesyl diphosphate specific) | | Authors: | Kurokawa, H, Ambo, T, Takahashi, S, Koyama, T. | | Deposit date: | 2020-08-07 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Thermobifida fusca cis-prenyltransferase reveals the dynamic nature of its RXG motif-mediated inter-subunit interactions critical for its catalytic activity.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
7CPN
 
 | | CRYSTAL STRUCTURE OF DODECAPRENYL DIPHOSPHATE SYNTHASE FROM THERMOBIFIDA FUSCA | | Descriptor: | GLYCEROL, SULFATE ION, Trans,polycis-polyprenyl diphosphate synthase ((2Z,6E)-farnesyl diphosphate specific) | | Authors: | Kurokawa, H, Ambo, T, Takahasi, S, Koyama, T. | | Deposit date: | 2020-08-07 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of Thermobifida fusca cis-prenyltransferase reveals the dynamic nature of its RXG motif-mediated inter-subunit interactions critical for its catalytic activity.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
1IQ5
 
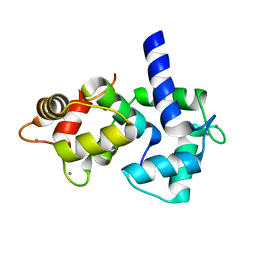 | | Calmodulin/nematode CA2+/Calmodulin dependent kinase kinase fragment | | Descriptor: | CA2+/CALMODULIN DEPENDENT KINASE KINASE, CALCIUM ION, CALMODULIN | | Authors: | Kurokawa, H, Osawa, M, Kurihara, H, Katayama, N, Tokumitsu, H, Swindells, M.B, Kainosho, M, Ikura, M. | | Deposit date: | 2001-06-14 | | Release date: | 2001-09-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Target-induced conformational adaptation of calmodulin revealed by the crystal structure of a complex with nematode Ca(2+)/calmodulin-dependent kinase kinase peptide
J.Mol.Biol., 312, 2001
|
|
1OVT
 
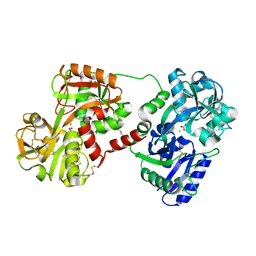 | |
1V9Y
 
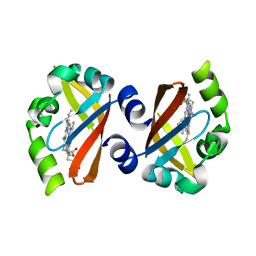 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (ferric form) | | Descriptor: | Heme pas sensor protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Lee, D.S, Watanabe, M, Sagami, I, Mikami, B, Raman, C.S, Shimizu, T. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor.
J.Biol.Chem., 279, 2004
|
|
1V9Z
 
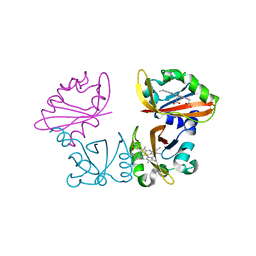 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (Ferrous Form) | | Descriptor: | Heme pas sensor protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Lee, D.S, Watanabe, M, Sagami, I, Mikami, B, Raman, C.S, Shimizu, T. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor.
J.Biol.Chem., 279, 2004
|
|
1VB6
 
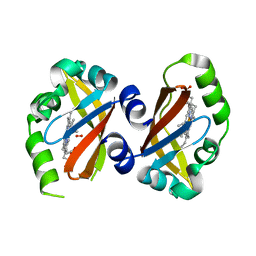 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (oxygen-bound form) | | Descriptor: | Heme pas sensor protein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Watanabe, M, Sagami, I, Mikami, B, Shimizu, T. | | Deposit date: | 2004-02-24 | | Release date: | 2005-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of oxygen-bound form of a Heme PAS domain of Ec DOS
To be Published
|
|
3ADE
 
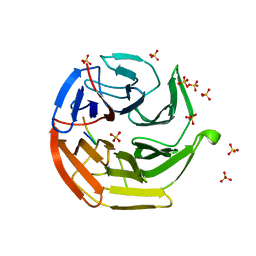 | | Crystal Structure of Keap1 in Complex with Sequestosome-1/p62 | | Descriptor: | Kelch-like ECH-associated protein 1, SULFATE ION, Sequestosome-1 | | Authors: | Kurokawa, H, Yamamoto, M. | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1
Nat.Cell Biol., 12, 2010
|
|
1NFT
 
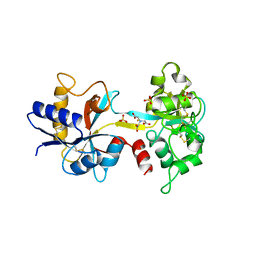 | | OVOTRANSFERRIN, N-TERMINAL LOBE, IRON LOADED OPEN FORM | | Descriptor: | FE (III) ION, NITRILOTRIACETIC ACID, PROTEIN (OVOTRANSFERRIN), ... | | Authors: | Mizutani, K, Yamashita, H, Kurokawa, H, Mikami, B, Hirose, M. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Alternative structural state of transferrin. The crystallographic analysis of iron-loaded but domain-opened ovotransferrin N-lobe.
J.Biol.Chem., 274, 1999
|
|
1J1R
 
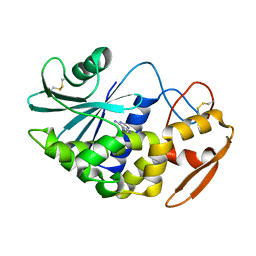 | | Structure of Pokeweed Antiviral Protein from Seeds (PAP-S1) Complexed with Adenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Antiviral Protein S | | Authors: | Watanabe, K, Sato, E, Honjo, E, Motoshima, H, Kurokawa, H, Mikami, B, Monzingo, A.F, Robertus, J.D, Fujii, H, Hidaka, A. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Pokweed Antiviral Protein from Seeds (PAP-S1) at 1.8 Angstrom Resolution
To be published
|
|
1J1Q
 
 | | Structure of Pokeweed Antiviral Protein from Seeds (PAP-S1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antiviral protein S | | Authors: | Watanabe, K, Sato, E, Honjo, E, Motoshima, H, Kurokawa, H, Mikami, B, Monzingo, A.F, Robertus, J.D, Fujii, H, Hidaka, A. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Pokweed Antiviral Protein from Seeds (PAP-S1) at 1.8 Angstrom Resolution
To be published
|
|
1J1S
 
 | | Pokeweed Antiviral Protein from Seeds (PAP-S1) Complexed with Formycin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antiviral Protein S, FORMYCIN-5'-MONOPHOSPHATE | | Authors: | Watanabe, K, Sato, E, Honjo, E, Motoshima, H, Kurokawa, H, Mikami, B, Monzingo, A.F, Robertus, J.D, Fujii, H, Hidaka, A. | | Deposit date: | 2002-12-14 | | Release date: | 2004-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Pokweed Antiviral Protein from Seeds (PAP-S1) at 1.8 Angstrom Resolution
To be published
|
|
1JOY
 
 | | SOLUTION STRUCTURE OF THE HOMODIMERIC DOMAIN OF ENVZ FROM ESCHERICHIA COLI BY MULTI-DIMENSIONAL NMR. | | Descriptor: | PROTEIN (ENVZ_ECOLI) | | Authors: | Tomomori, C, Tanaka, T, Dutta, R, Park, H, Saha, S.K, Zhu, Y, Ishima, R, Liu, D, Tong, K.I, Kurokawa, H, Qian, H, Inouye, M, Ikura, M. | | Deposit date: | 1998-12-28 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ.
Nat.Struct.Biol., 6, 1999
|
|
8ILL
 
 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH5.6 | | Descriptor: | CHLORIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, green fluorescent protein | | Authors: | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
8ILK
 
 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH8.5 | | Descriptor: | CHLORIDE ION, Green FLUORESCENT PROTEIN | | Authors: | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
1TFA
 
 | | OVOTRANSFERRIN, N-TERMINAL LOBE, APO FORM | | Descriptor: | PROTEIN (OVOTRANSFERRIN), SULFATE ION | | Authors: | Mizutani, K, Yamashita, H, Mikami, B, Hirose, M. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternative structural state of transferrin. The crystallographic analysis of iron-loaded but domain-opened ovotransferrin N-lobe.
J.Biol.Chem., 274, 1999
|
|
