2LIE
 
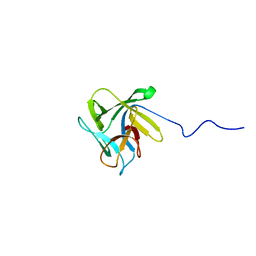 | | NMR structure of the lectin CCL2 | | Descriptor: | CCL2 lectin | | Authors: | Schubert, M, Walti, M.A, Egloff, P, Bleuler-Martinez, S, Aebi, M, Allain, F.F.-H, Kunzler, M. | | Deposit date: | 2011-08-29 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System
Plos Pathog., 8, 2012
|
|
4USP
 
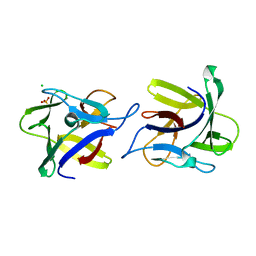 | | X-ray structure of the dimeric CCL2 lectin in native form | | Descriptor: | CCL2 LECTIN, CHLORIDE ION, PHOSPHATE ION | | Authors: | Bleuler-Martinez, S, Varrot, A, Schubert, M, Stutz, M, Sieber, R, Hengartner, M, Aebi, M, Kunzler, M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dimerization of the fungal defense lectin CCL2 is essential for its toxicity against nematodes.
Glycobiology, 27, 2017
|
|
4USO
 
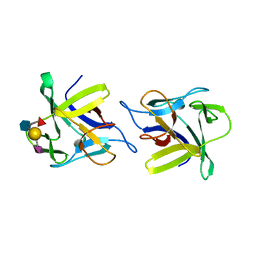 | | X-ray structure of the CCL2 lectin in complex with sialyl lewis X | | Descriptor: | CCL2 LECTIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bleuler-Martinez, S, Varrot, A, Schubert, M, Stutz, M, Sieber, R, Hengartner, M, Aebi, M, Kunzler, M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dimerization of the fungal defense lectin CCL2 is essential for its toxicity against nematodes.
Glycobiology, 27, 2017
|
|
2JJ6
 
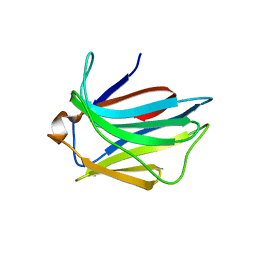 | | Crystal structure of the putative carbohydrate recognition domain of the human galectin-related protein | | Descriptor: | GALECTIN-RELATED PROTEIN | | Authors: | Thore, S, Walti, M.A, Kunzler, M, Aebi, M. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Putative Carbohydrate Recognition Domain of Human Galectin-Related Protein
Proteins: Struct., Funct., Bioinf., 72, 2008
|
|
5FSB
 
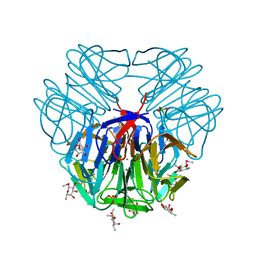 | | Structure of tectonin 2 from laccaria bicolor in complex with 2-o-methyl-methyl-seleno-beta-l-fucopyranoside | | Descriptor: | BORIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Sommer, R, Bleuer, S, Titz, A, Kunzler, M, Varrot, A. | | Deposit date: | 2016-01-04 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of Fungal Tectonin in Complex with O-Methylated Glycans Suggest Key Role in Innate Immune Defense.
Structure, 26, 2018
|
|
5FSC
 
 | | Structure of tectonin 2 from Laccaria bicolor in complex with allyl- alpha_4-methyl-mannoside | | Descriptor: | MAGNESIUM ION, TECTONIN 2, prop-2-en-1-yl 4-O-methyl-alpha-D-mannopyranoside | | Authors: | Sommer, R, Bleuer, S, Titz, A, Kunzler, M, Varrot, A. | | Deposit date: | 2016-01-04 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Fungal Tectonin in Complex with O-Methylated Glycans Suggest Key Role in Innate Immune Defense.
Structure, 26, 2018
|
|
2LIQ
 
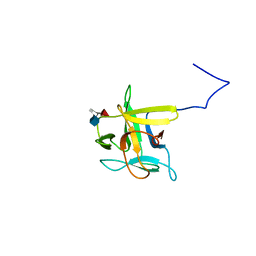 | | Solution structure of CCL2 in complex with glycan | | Descriptor: | CCL2 lectin, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Schubert, M, Bleuler-Martinez, S, Walti, M.A, Egloff, P, Aebi, M, Kuenzler, M, Allain, F.H.-T. | | Deposit date: | 2011-08-30 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System
Plos Pathog., 8, 2012
|
|
7ZNX
 
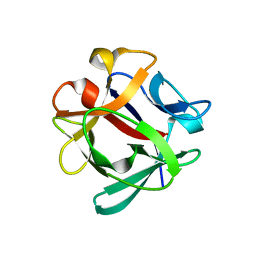 | |
2WKK
 
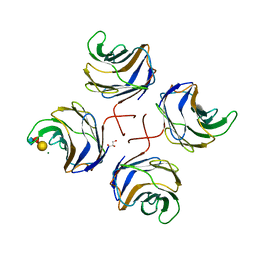 | | Identification of the glycan target of the nematotoxic fungal galectin CGL2 in Caenorhabditis elegans | | Descriptor: | GALECTIN-2, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Butschi, A, Titz, A, Waelti, M, Olieric, V, Paschinger, K, Xiaoqiang, G, Seeberger, P.H, Wilson, I.B.H, Aebi, M, Hengartner, M.O, Kuenzler, M. | | Deposit date: | 2009-06-14 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Caenorhabditis Elegans N-Glycan Core Beta-Galactoside Confers Sensitivity Towards Nematotoxic Fungal Galectin Cgl2.
Plos Pathog., 6, 2010
|
|
6GEW
 
 | | OphA Y63F-sinefungin complex | | Descriptor: | OphA, S-ADENOSYL-L-HOMOCYSTEINE, SINEFUNGIN | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2018-04-27 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A molecular mechanism for the enzymatic methylation of nitrogen atoms within peptide bonds.
Sci Adv, 4, 2018
|
|
2MN5
 
 | | NMR structure of Copsin | | Descriptor: | Copsin | | Authors: | Hofmann, D, Wider, G, Essig, A, Aebi, M. | | Deposit date: | 2014-03-28 | | Release date: | 2014-10-29 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Copsin, a Novel Peptide-based Fungal Antibiotic Interfering with the Peptidoglycan Synthesis.
J.Biol.Chem., 289, 2014
|
|
5N0N
 
 | |
5N0U
 
 | |
5N0P
 
 | | Crystal structure of OphA-DeltaC18 in complex with SAH | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, peptide N-methyltranferase | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A molecular mechanism for the enzymatic methylation of nitrogen atoms within peptide bonds.
Sci Adv, 4, 2018
|
|
5N0X
 
 | | Crystal structure of OphA-DeltaC6 in complex with SAM | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Peptide N-Methyltransferase, ... | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A molecular mechanism for the enzymatic methylation of nitrogen atoms within peptide bonds.
Sci Adv, 4, 2018
|
|
5N0S
 
 | |
5N0T
 
 | |
5N0R
 
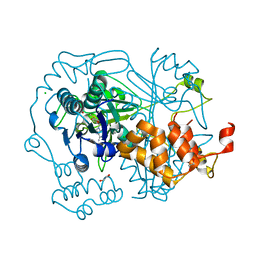 | |
6QZY
 
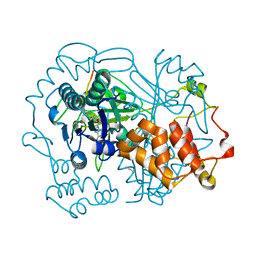 | | full length OphA V406P in complex with SAH | | Descriptor: | ASN-GLY-PHE-PRO-TRP-MVA-ILE-MVA-VAL-GLY-PRO-ILE-GLY, MAGNESIUM ION, Peptide N-methyltransferase, ... | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Substrate Plasticity of a Fungal Peptide alpha-N-Methyltransferase.
Acs Chem.Biol., 15, 2020
|
|
6QZZ
 
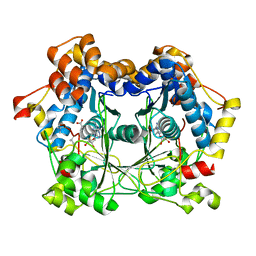 | | full length OphA V404E in complex with SAH | | Descriptor: | Peptide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Plasticity of a Fungal Peptide alpha-N-Methyltransferase.
Acs Chem.Biol., 15, 2020
|
|
6R00
 
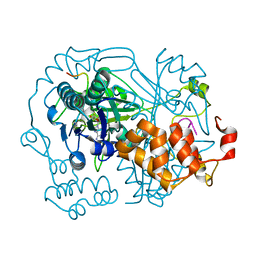 | | OphA DeltaC6 V404F complex with SAH | | Descriptor: | PHE-PRO-TRP-MVA-ILE-MVA-PHE-GLY-VAL-ILE-GLY-VAL-ILE-GLY, Peptide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Substrate Plasticity of a Fungal Peptide alpha-N-Methyltransferase.
Acs Chem.Biol., 15, 2020
|
|
6ZRW
 
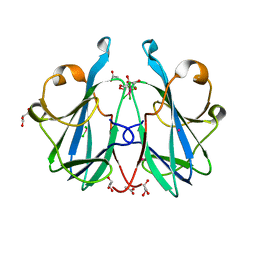 | | Crystal structure of the fungal lectin CML1 | | Descriptor: | ACETATE ION, GLYCEROL, Mucin-binding lectin 1, ... | | Authors: | Bleuler-Martinez, S, Olieric, V, Sharpe, M, Capitani, G, Aebi, M, Kuenzler, M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-function relationship of a novel fucoside-binding fruiting body lectin from Coprinopsis cinerea exhibiting nematotoxic activity.
Glycobiology, 32, 2022
|
|
7ZAZ
 
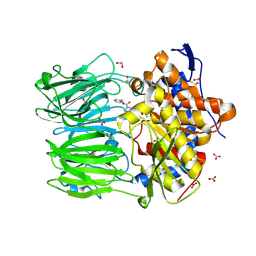 | | macrocyclase OphP with ZPP | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2022-03-23 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Biorxiv, 2022
|
|
7ZB2
 
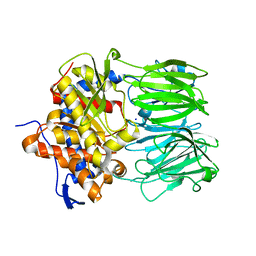 | | apo macrocyclase OphP | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2022-03-23 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Biorxiv, 2022
|
|
7ZB1
 
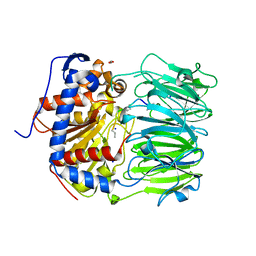 | | S580A with 18mer | | Descriptor: | 1,2-ETHANEDIOL, 18mer, BICARBONATE ION, ... | | Authors: | Song, H, Naismith, J.H. | | Deposit date: | 2022-03-23 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Biorxiv, 2022
|
|
