6OJW
 
 | | Crystal structure of Sphingomonas paucimobilis TMY1009 holo-LsdA | | Descriptor: | FE (III) ION, GLYCEROL, Lignostilbene-alpha,beta-dioxygenase isozyme I, ... | | Authors: | Kuatsjah, E, Verstraete, M.M, Kobylarz, M.J, Liu, A.K.N, Murphy, M.E.P, Eltis, L.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase.
J.Biol.Chem., 294, 2019
|
|
5VN5
 
 | | Crystal structure of LigY from Sphingobium sp. strain SYK-6 | | Descriptor: | 2,2',3-trihydroxy-3'-methoxy-5,5'-dicarboxybiphenyl meta-cleavage compound hydrolase, CHLORIDE ION, ZINC ION | | Authors: | Kuatsjah, E, Chan, A.C.K, Kobylarz, M.J, Murphy, M.E.P, Eltis, L.D. | | Deposit date: | 2017-04-28 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The bacterialmeta-cleavage hydrolase LigY belongs to the amidohydrolase superfamily, not to the alpha / beta-hydrolase superfamily.
J. Biol. Chem., 292, 2017
|
|
6OJT
 
 | | Crystal structure of Sphingomonas paucimobilis TMY1009 LsdA phenylazophenol complex | | Descriptor: | 4-Hydroxyazobenzene, FE (III) ION, Lignostilbene-alpha,beta-dioxygenase isozyme I | | Authors: | Kuatsjah, E, Verstraete, M.M, Kobylarz, M.J, Liu, A.K.N, Murphy, M.E.P, Eltis, L.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase.
J.Biol.Chem., 294, 2019
|
|
6OJR
 
 | | Crystal structure of Sphingomonas paucimobilis TMY1009 apo-LsdA | | Descriptor: | GLYCEROL, Lignostilbene-alpha,beta-dioxygenase isozyme I, MAGNESIUM ION | | Authors: | Kuatsjah, E, Verstraete, M.M, Kobylarz, M.J, Liu, A.K.N, Murphy, M.E.P, Eltis, L.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of functionally important residues and structural features in a bacterial lignostilbene dioxygenase.
J.Biol.Chem., 294, 2019
|
|
6XM9
 
 | | Crystal structure of vanillin bound to Co-LSD4 from Sphingobium sp. strain SYK-6 | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, ACETATE ION, COBALT (II) ION, ... | | Authors: | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2020-06-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
6XM7
 
 | | Crystal structure of DCA-S bound to Co-LSD4 from Sphingobium sp. strain SYK-6 | | Descriptor: | (2E)-3-{4-hydroxy-3-[(E)-2-(4-hydroxy-3-methoxyphenyl)ethenyl]-5-methoxyphenyl}prop-2-enoic acid, COBALT (II) ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2020-06-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
6XM6
 
 | | Crystal structure of cobalt-bound LSD4 from Sphingobium sp. strain SYK-6 | | Descriptor: | COBALT (II) ION, Dioxygenase | | Authors: | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2020-06-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
6XM8
 
 | | Crystal structure of lignostilbene bound to Co-LSD4 from Sphingobium sp. strain SYK-6 | | Descriptor: | COBALT (II) ION, DIMETHYL SULFOXIDE, Dioxygenase, ... | | Authors: | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2020-06-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
6XMA
 
 | | Crystal structure of iron-bound LSD4 from Sphingobium sp. strain SYK-6 | | Descriptor: | Dioxygenase, FE (III) ION, SULFATE ION | | Authors: | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2020-06-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
6E6I
 
 | | Crystal structure of 4-methyl HOPDA bound to LigY from Sphingobium sp. strain SYK-6 | | Descriptor: | (1Z,3E)-5-carboxy-3-methyl-5-oxo-1-phenylpenta-1,3-dien-1-olate, 2,2',3-trihydroxy-3'-methoxy-5,5'-dicarboxybiphenyl meta-cleavage compound hydrolase, ZINC ION | | Authors: | Kuatsjah, E, Chan, A.C, Hurst, T.E, Snieckus, V, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2018-07-24 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal- and Serine-Dependent Meta-Cleavage Product Hydrolases Utilize Similar Nucleophile-Activation Strategies
Acs Catalysis, 8, 2018
|
|
8ABV
 
 | | Crystal structure of SpLdpA in complex with erythro-DGPD | | Descriptor: | (1R,2S)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, (1S,2R)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, GLYCEROL, ... | | Authors: | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ABT
 
 | | Crystal structure of NaLdpA in complex with the product analog Resveratrol | | Descriptor: | RESVERATROL, SULFATE ION, SnoaL-like domain-containing protein | | Authors: | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ABU
 
 | | Crystal structure of NaLdpA mutant H97Q in complex with erythro-DGPD | | Descriptor: | (1S,2R)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, SnoaL-like domain-containing protein | | Authors: | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ABW
 
 | | Crystal structure of SpLdpA in complex with threo-DGPD | | Descriptor: | (1R,2R)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, (1S,2S)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, SULFATE ION, ... | | Authors: | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8AYV
 
 | |
7ON9
 
 | | Crystal structure of para-hydroxybenzoate-3-hydroxylase PraI | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID | | Authors: | Zahn, M, McGeehan, J.E. | | Deposit date: | 2021-05-25 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Debottlenecking 4-hydroxybenzoate hydroxylation in Pseudomonas putida KT2440 improves muconate productivity from p-coumarate.
Metab Eng, 70, 2022
|
|
7N79
 
 | | O2-, PLP-dependent desaturase Plu4 holo-enzyme | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Hoffarth, E.R, Ryan, K.S. | | Deposit date: | 2021-06-09 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A shared mechanistic pathway for pyridoxal phosphate-dependent arginine oxidases.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7FHR
 
 | | Crystal Structure of a Rieske Oxygenase from Cupriavidus metallidurans | | Descriptor: | 1,2-ETHANEDIOL, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Mahto, J.K, Dhankhar, P, Kumar, P. | | Deposit date: | 2021-07-30 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by phthalate dioxygenase from Comamonas testosteroni.
J.Biol.Chem., 297, 2021
|
|
7FJL
 
 | |
7V25
 
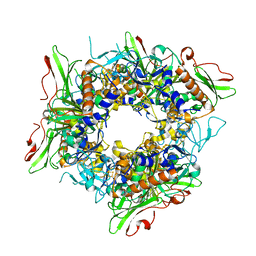 | |
7V28
 
 | |
7QWT
 
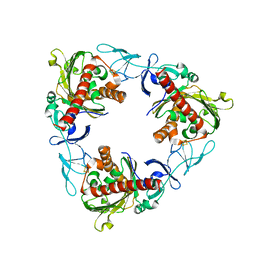 | | Rieske non-heme iron monooxygenase for guaiacol O-demethylation | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Rieske (2Fe-2S) domain protein | | Authors: | Hinchen, D.J, Zahn, M, Bleem, A, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.712 Å) | | Cite: | Discovery, characterization, and metabolic engineering of Rieske non-heme iron monooxygenases for guaiacol O-demethylation
Chem Catal, 2022
|
|
7RGB
 
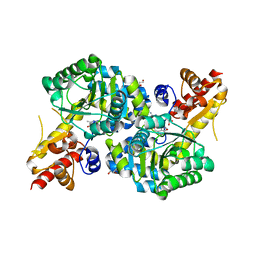 | | O2-, PLP-dependent desaturase Plu4 product-bound enzyme | | Descriptor: | (2Z,4E)-5-carbamimidamido-2-iminopent-4-enoic acid, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Hoffarth, E.R, Ryan, K.S. | | Deposit date: | 2021-07-14 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A shared mechanistic pathway for pyridoxal phosphate-dependent arginine oxidases.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RF9
 
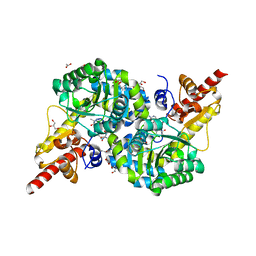 | | O2-, PLP-dependent desaturase Plu4 intermediate-bound enzyme | | Descriptor: | (2E)-5-carbamimidamido-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}pentanoic acid, 1,2-ETHANEDIOL, 2-(2-ETHOXYETHOXY)ETHANOL, ... | | Authors: | Hoffarth, E.R, Ryan, K.S. | | Deposit date: | 2021-07-13 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | A shared mechanistic pathway for pyridoxal phosphate-dependent arginine oxidases.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6C3D
 
 | | O2-, PLP-dependent L-arginine hydroxylase RohP quinonoid II complex | | Descriptor: | (2E,3E)-5-carbamimidamido-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}pent-3-enoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hedges, J.B, Ryan, K.S. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Snapshots of the Catalytic Cycle of an O2, Pyridoxal Phosphate-Dependent Hydroxylase.
ACS Chem. Biol., 13, 2018
|
|
