7UMH
 
 | | Energetic robustness to large scale structural dynamics in a photosynthetic supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Harris, D, Toporik, H, Schlau-Cohen, G.S, Mazor, Y. | | Deposit date: | 2022-04-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Energetic robustness to large scale structural fluctuations in a photosynthetic supercomplex.
Nat Commun, 14, 2023
|
|
6FEJ
 
 | | Anabaena Apo-C-Terminal Domain Homolog Protein | | Descriptor: | All4940 protein, UREA | | Authors: | Harris, D, Wilson, A, Muzzopappa, F, Kirilovsky, D, Adir, N. | | Deposit date: | 2018-01-02 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural rearrangements in the C-terminal domain homolog of Orange Carotenoid Protein are crucial for carotenoid transfer.
Commun Biol, 1, 2018
|
|
6S5L
 
 | |
2JD4
 
 | | Mouse laminin alpha1 chain, domains LG4-5 | | Descriptor: | CHLORIDE ION, LAMININ SUBUNIT ALPHA-1, MAGNESIUM ION | | Authors: | Harrison, D, Hussain, S.A, Combs, A.C, Ervasti, J.M, Yurchenco, P.D, Hohenester, E. | | Deposit date: | 2007-01-04 | | Release date: | 2007-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Cell Surface Anchorage Sites of Laminin {Alpha}1Lg4-5.
J.Biol.Chem., 282, 2007
|
|
8FB9
 
 | | LH2-LH3 antenna in anti parallel configuration embedded in a nanodisc | | Descriptor: | BACTERIOCHLOROPHYLL A, LYCOPENE, Light-harvesting protein B-800/850 alpha chain, ... | | Authors: | Toporik, H, Harris, D, Schlau-Cohen, G.S, Mazor, Y. | | Deposit date: | 2022-11-29 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Elucidating interprotein energy transfer dynamics within the antenna network from purple bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FBB
 
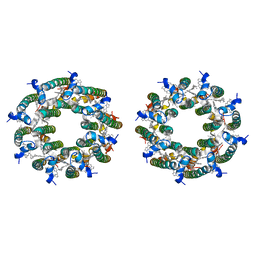 | | LH2-LH3 antenna in parallel configuration embedded in a nanodisc | | Descriptor: | BACTERIOCHLOROPHYLL A, LYCOPENE, Light-harvesting protein B-800/850 alpha chain, ... | | Authors: | Toporik, H, Harris, D, Schlau-Cohen, G.S, Mazor, Y. | | Deposit date: | 2022-11-29 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | Elucidating interprotein energy transfer dynamics within the antenna network from purple bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7TV3
 
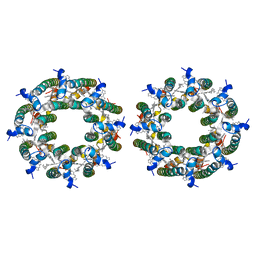 | | LH2-LH3 antenna in parallel configuration embedded in a nanodisc | | Descriptor: | BACTERIOCHLOROPHYLL A, LYCOPENE, Light-harvesting protein B-800/850 alpha chain, ... | | Authors: | Toporik, H, Harris, D, Schlau-Cohen, G.S, Mazor, Y. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (11.4 Å) | | Cite: | Elucidating interprotein energy transfer dynamics within the antenna network from purple bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7TUW
 
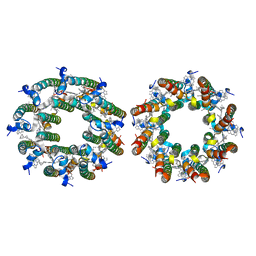 | | LH2-LH3 antenna in anti parallel configuration embedded in a nanodisc | | Descriptor: | BACTERIOCHLOROPHYLL A, LYCOPENE, Light-harvesting protein B-800/850 alpha chain, ... | | Authors: | Toporik, H, Harris, D, Schlau-Cohen, G.S, Mazor, Y. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Elucidating interprotein energy transfer dynamics within the antenna network from purple bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1HTT
 
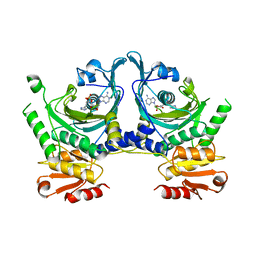 | | HISTIDYL-TRNA SYNTHETASE | | Descriptor: | ADENOSINE MONOPHOSPHATE, HISTIDINE, HISTIDYL-TRNA SYNTHETASE | | Authors: | Arnez, J.G, Harris, D.C, Mitschler, A, Rees, B, Francklyn, C.S, Moras, D. | | Deposit date: | 1996-03-09 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of histidyl-tRNA synthetase from Escherichia coli complexed with histidyl-adenylate.
EMBO J., 14, 1995
|
|
5OOK
 
 | | Structure of A. marina Phycocyanin contains overlapping isoforms | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHYCOCYANOBILIN, Phycocyanin, ... | | Authors: | Bar-Zvi, S, Lahav, A, Blankenship, E.R, Adir, N. | | Deposit date: | 2017-08-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural heterogeneity leads to functional homogeneity in A. marina phycocyanin.
Biochim. Biophys. Acta, 1859, 2018
|
|
1KMN
 
 | |
1KMM
 
 | |
6B9C
 
 | | Superfolder Green Fluorescent Protein with 4-nitro-L-phenylalanine at the chromophore (position 66) | | Descriptor: | CARBON DIOXIDE, Green fluorescent protein | | Authors: | Phillips-Piro, C.M, Brewer, S.H, Olenginski, G.M, Piacentini, J. | | Deposit date: | 2017-10-10 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Structural and spectrophotometric investigation of two unnatural amino-acid altered chromophores in the superfolder green fluorescent protein
Acta Crystallogr.,Sect.D, 2021
|
|
8TQD
 
 | | NF-Kappa-B1 Bound with a Covalent Inhibitor | | Descriptor: | 1-(2-bromo-4-chlorophenyl)-N-{(3S)-1-[(E)-iminomethyl]pyrrolidin-3-yl}methanesulfonamide, Nuclear factor NF-kappa-B p105 subunit | | Authors: | Hilbert, B.J. | | Deposit date: | 2023-08-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | DrugMap: A quantitative pan-cancer analysis of cysteine ligandability.
Cell, 187, 2024
|
|
3UPU
 
 | | Crystal structure of the T4 Phage SF1B Helicase Dda | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*T)-3', ATP-dependent DNA helicase dda | | Authors: | He, X, Yun, M.K, Pemble IV, C.W, Kreuzer, K.N, Raney, K.D, White, S.W. | | Deposit date: | 2011-11-18 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | The T4 Phage SF1B Helicase Dda Is Structurally Optimized to Perform DNA Strand Separation.
Structure, 20, 2012
|
|
5V1B
 
 | | Structure of PHD1 in complex with 1,2,4-Triazolo-[1,5-a]pyridine | | Descriptor: | 4-([1,2,4]triazolo[1,5-a]pyridin-5-yl)benzonitrile, Egl nine homolog 2, FE (III) ION, ... | | Authors: | Skene, R.J. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | 1,2,4-Triazolo-[1,5-a]pyridine HIF Prolylhydroxylase Domain-1 (PHD-1) Inhibitors With a Novel Monodentate Binding Interaction.
J. Med. Chem., 60, 2017
|
|
5V18
 
 | | Structure of PHD2 in complex with 1,2,4-Triazolo-[1,5-a]pyridine | | Descriptor: | 4-([1,2,4]triazolo[1,5-a]pyridin-5-yl)benzonitrile, Egl nine homolog 1, FE (II) ION, ... | | Authors: | Skene, R.J. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 1,2,4-Triazolo-[1,5-a]pyridine HIF Prolylhydroxylase Domain-1 (PHD-1) Inhibitors With a Novel Monodentate Binding Interaction.
J. Med. Chem., 60, 2017
|
|
1CRL
 
 | |
1THG
 
 | | 1.8 ANGSTROMS REFINED STRUCTURE OF THE LIPASE FROM GEOTRICHUM CANDIDUM | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schrag, J.D, Cygler, M. | | Deposit date: | 1992-07-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 A refined structure of the lipase from Geotrichum candidum.
J.Mol.Biol., 230, 1993
|
|
1LPS
 
 | | A STRUCTURAL BASIS FOR THE CHIRAL PREFERENCES OF LIPASES | | Descriptor: | (1S)-MENTHYL HEXYL PHOSPHONATE GROUP, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grochulski, P.G, Cygler, M.C. | | Deposit date: | 1995-01-05 | | Release date: | 1995-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A Structural Basis for the Chiral Preferences of Lipases
J.Am.Chem.Soc., 116, 1994
|
|
1LPP
 
 | |
1LPO
 
 | |
1LPM
 
 | | A STRUCTURAL BASIS FOR THE CHIRAL PREFERENCES OF LIPASES | | Descriptor: | (1R)-MENTHYL HEXYL PHOSPHONATE GROUP, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grochulski, P.G, Cygler, M.C. | | Deposit date: | 1995-01-06 | | Release date: | 1995-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A Structural Basis for the Chiral Preferences of Lipases
J.Am.Chem.Soc., 116, 1994
|
|
1LPN
 
 | |
1TRH
 
 | |
