1N8M
 
 | | Solution structure of Pi4, a four disulfide bridged scorpion toxin active on potassium channels | | Descriptor: | Potassium channel blocking toxin 4 | | Authors: | Guijarro, J.I, M'Barek, S, Olamendi-Portugal, T, Gomez-Lagunas, F, Garnier, D, Rochat, H, Possani, L.D, Sabatier, J.M, Delepierre, M. | | Deposit date: | 2002-11-21 | | Release date: | 2003-09-02 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pi4, a short four-disulfide-bridged scorpion toxin specific of potassium channels.
Protein Sci., 12, 2003
|
|
1JH3
 
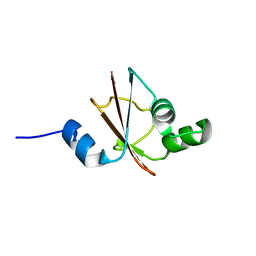 | | Solution structure of tyrosyl-tRNA synthetase C-terminal domain. | | Descriptor: | TYROSYL-TRNA SYNTHETASE | | Authors: | Guijarro, J.I, Pintar, A, Prochnicka-Chalufour, A, Guez, V, Gilquin, B, Bedouelle, H, Delepierre, M. | | Deposit date: | 2001-06-27 | | Release date: | 2002-03-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Anticodon Arm Binding Domain of Bacillus stearothermophilus
Tyrosyl-tRNA Synthetase
Structure, 10, 2002
|
|
7ZRU
 
 | |
6GCJ
 
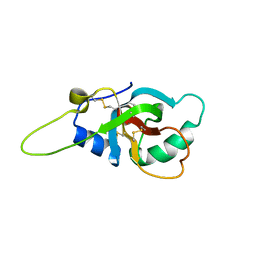 | | Solution structure of the RodA hydrophobin from Aspergillus fumigatus | | Descriptor: | Hydrophobin | | Authors: | Pille, A, Kwan, A, Aimanianda, V, Latge, J.-P, Sunde, M, Guijarro, J.I. | | Deposit date: | 2018-04-18 | | Release date: | 2019-03-27 | | Last modified: | 2019-09-04 | | Method: | SOLUTION NMR | | Cite: | Assembly and disassembly of Aspergillus fumigatus conidial rodlets
Cell Surf, 2019
|
|
6NW8
 
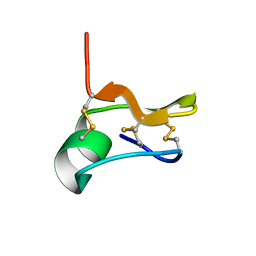 | | SOLUTION STRUCTURE OF CN29, A TOXIN FROM CENTRUROIDES NOXIUS SCORPION VENOM | | Descriptor: | Cn29 | | Authors: | Delepierre, M, Gurrola, G.B, Possani, L.D, Guijarro, J.I. | | Deposit date: | 2019-02-06 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Cn29, a novel orphan peptide found in the venom of the scorpion Centruroides noxius: Structure and function.
Toxicon, 167, 2019
|
|
2KEL
 
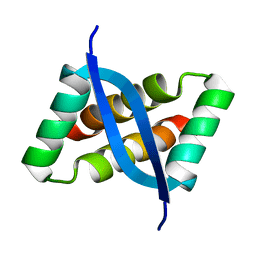 | | Structure of the transcription regulator SvtR from the hyperthermophilic archaeal virus SIRV1 | | Descriptor: | Uncharacterized protein 56B | | Authors: | Guilliere, F, Kessler, A, Peixeiro, N, Sezonov, G, Prangishvili, D, Delepierre, M, Guijarro, J.I. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, function, and targets of the transcriptional regulator SvtR from the hyperthermophilic archaeal virus SIRV1.
J.Biol.Chem., 284, 2009
|
|
2LVH
 
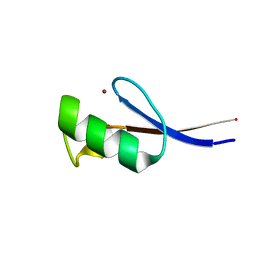 | | Solution structure of the zinc finger AFV1p06 protein from the hyperthermophilic archaeal virus AFV1 | | Descriptor: | Putative zinc finger protein ORF59a, ZINC ION | | Authors: | Guilliere, F, Sezonov, G, Prangishvili, D, Delepierre, M, Guijarro, J. | | Deposit date: | 2012-07-05 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an archaeal DNA binding protein with an eukaryotic zinc finger fold.
Plos One, 8, 2013
|
|
7PSZ
 
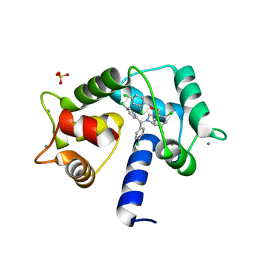 | | Crystal structure of CaM in complex with CDZ (form 1) | | Descriptor: | 1-[bis(4-chlorophenyl)methyl]-3-[(2~{R})-2-(2,4-dichlorophenyl)-2-[(2,4-dichlorophenyl)methoxy]ethyl]imidazole, CALCIUM ION, Calmodulin-1, ... | | Authors: | Mechaly, A.E, Leger, C, Haouz, A, Chenal, A. | | Deposit date: | 2021-09-24 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium.
Bmc Biol., 20, 2022
|
|
7PU9
 
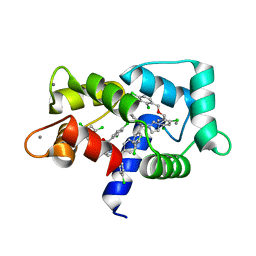 | | Crystal structure of CaM in complex with CDZ (form 2) | | Descriptor: | 1-[bis(4-chlorophenyl)methyl]-3-[(2~{R})-2-(2,4-dichlorophenyl)-2-[(2,4-dichlorophenyl)methoxy]ethyl]imidazole, CALCIUM ION, Calmodulin-1 | | Authors: | Mechaly, A.E, Leger, C, Haouz, A, Chenal, A. | | Deposit date: | 2021-09-28 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.279 Å) | | Cite: | Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium.
Bmc Biol., 20, 2022
|
|
