4PM3
 
 | |
4EC6
 
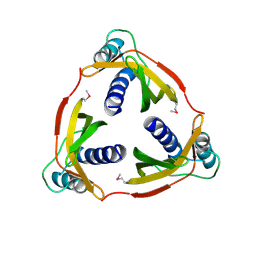 | | Ntf2-like, potential transfer protein TraM from Gram-positive conjugative plasmid pIP501 | | Descriptor: | Putative uncharacterized protein | | Authors: | Goessweiner-Mohr, N, Grumet, L, Pavkov-Keller, T, Wang, M, Keller, W. | | Deposit date: | 2012-03-26 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 A Structure of the Enterococcus Conjugation Protein TraM resembles VirB8 Type IV Secretion Proteins.
J.Biol.Chem., 288, 2013
|
|
4HIC
 
 | |
4P0Z
 
 | |
4P0Y
 
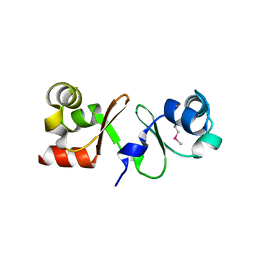 | |
6G1T
 
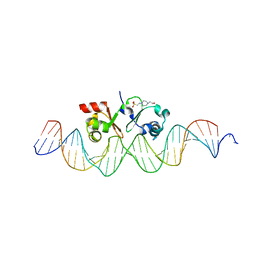 | | TraN, a repressor of an Enterococcus conjugative type IV secretion system | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AM32, DNA (34-MER) | | Authors: | Goessweiner-Mohr, N, Kohler, V, Keller, W. | | Deposit date: | 2018-03-22 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | TraN: A novel repressor of an Enterococcus conjugative type IV secretion system.
Nucleic Acids Res., 46, 2018
|
|
7AGX
 
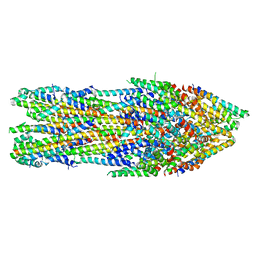 | | Apo-state type 3 secretion system export apparatus complex from Salmonella enterica typhimurium | | Descriptor: | Protein PrgI, Protein PrgJ, Surface presentation of antigens protein SpaP, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Miletic, S, Wald, J, Marlovits, T. | | Deposit date: | 2020-09-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation.
Nat Commun, 12, 2021
|
|
7PBU
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvA-HJ core [t2 dataset] | | Descriptor: | Holliday junction, Holliday junction ATP-dependent DNA helicase RuvA | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBQ
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0+A [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBS
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0+A [t1 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBR
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0-A [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB, MAGNESIUM ION, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
6SK7
 
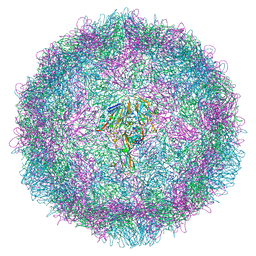 | | Cryo-EM structure of rhinovirus-A89 | | Descriptor: | VP1 capsid protein, VP2 capsid protein, VP3 capsid protein, ... | | Authors: | Wald, J, Goessweiner-Mohr, N, Blaas, D, Pasin, M. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of pleconaril-resistant rhinovirus-B5 complexed to the antiviral OBR-5-340 reveals unexpected binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6SK5
 
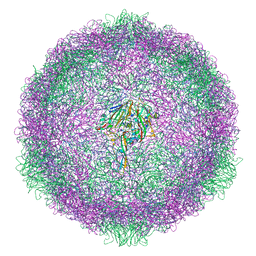 | | Cryo-EM structure of rhinovirus-B5 complexed to antiviral OBR-5-340 | | Descriptor: | 6-phenyl-~{N}3-[4-(trifluoromethyl)phenyl]-1~{H}-pyrazolo[3,4-d]pyrimidine-3,4-diamine, Rhinovirus B5 VP1, Rhinovirus B5 VP2, ... | | Authors: | Wald, J, Goessweiner-Mohr, N, Blaas, D, Pasin, M. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of pleconaril-resistant rhinovirus-B5 complexed to the antiviral OBR-5-340 reveals unexpected binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6SK6
 
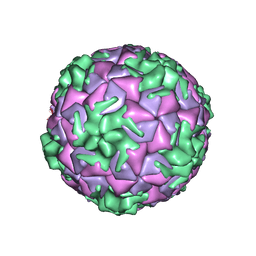 | | Cryo-EM structure of rhinovirus-B5 | | Descriptor: | Rhinovirus B5 VP1, Rhinovirus B5 VP2, Rhinovirus B5 VP3, ... | | Authors: | Wald, J, Goessweiner-Mohr, N, Blaas, D, Pasin, M. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of pleconaril-resistant rhinovirus-B5 complexed to the antiviral OBR-5-340 reveals unexpected binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7AHI
 
 | | Substrate-engaged type 3 secretion system needle complex from Salmonella enterica typhimurium - SpaR state 2 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, LAURYL DIMETHYLAMINE-N-OXIDE, Lipoprotein PrgK, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Miletic, S, Wald, J, Marlovits, T. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation
Nat Commun, 12, 2021
|
|
7AH9
 
 | | Substrate-engaged type 3 secretion system needle complex from Salmonella enterica typhimurium - SpaR state 1 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, LAURYL DIMETHYLAMINE-N-OXIDE, Lipoprotein PrgK, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Miletic, S, Wald, J, Marlovits, T. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation
Nat Commun, 12, 2021
|
|
7PBL
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s1 [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBM
 
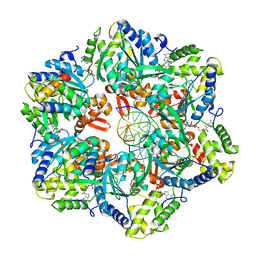 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s2 [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBO
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s4 [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBT
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s1 [t1 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Wald, J, Fahrenkamp, D, Goessweiner-Mohr, N, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBP
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s5 [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBN
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s3 [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
5AIW
 
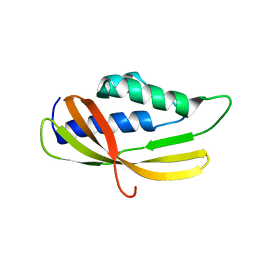 | |
