3KAJ
 
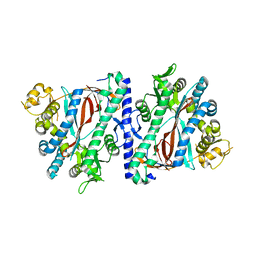 | | Apoenzyme structure of homoglutathione synthetase from Glycine max in open conformation | | Descriptor: | Homoglutathione synthetase | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
3KAK
 
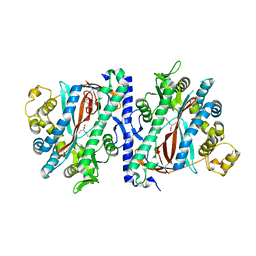 | | Structure of homoglutathione synthetase from Glycine max in open conformation with gamma-glutamyl-cysteine bound. | | Descriptor: | GAMMA-GLUTAMYLCYSTEINE, Homoglutathione synthetase | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
3KAL
 
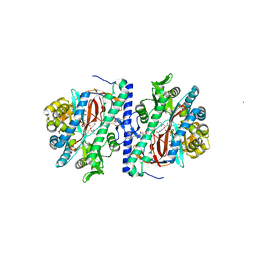 | | Structure of homoglutathione synthetase from Glycine max in closed conformation with homoglutathione, ADP, a sulfate ion, and three magnesium ions bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-gamma-glutamyl-L-cysteinyl-beta-alanine, MAGNESIUM ION, ... | | Authors: | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
3R8W
 
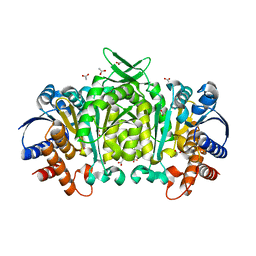 | | Structure of 3-isopropylmalate dehydrogenase isoform 2 from Arabidopsis thaliana at 2.2 angstrom resolution | | Descriptor: | 3-isopropylmalate dehydrogenase 2, chloroplastic, ACETATE ION | | Authors: | He, Y, Galant, A, Pang, Q, Strul, J.M, Balogun, S, Jez, J.M, Chen, S. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional evolution of isopropylmalate dehydrogenases in the leucine and glucosinolate pathways of Arabidopsis thaliana.
J.Biol.Chem., 286, 2011
|
|
