1SEF
 
 | |
1SFN
 
 | |
2NQ5
 
 | | Crystal structure of methyltransferase from Streptococcus mutans | | Descriptor: | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-30 | | Release date: | 2006-11-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of methyltransferase from Streptococcus mutans
To be Published
|
|
3V5C
 
 | | Crystal structure of the mutant E234A of Galacturonate Dehydratase from GEOBACILLUS SP. complexed with Mg | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Groninger-Poe, F, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-12-16 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of the mutant E234A of Galacturonate Dehydratase from GEOBACILLUS SP. complexed with Mg
To be Published
|
|
4O8R
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with 5,6-dihydrouridine 5'-monophosphate | | Descriptor: | 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, CHLORIDE ION, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-12-29 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with 5,6-dihydrouridine 5'-monophosphate
To be Published
|
|
4Z18
 
 | | CRYSTAL STRUCTURE OF HUMAN PD-L1 | | Descriptor: | CHLORIDE ION, Programmed cell death 1 ligand 1 | | Authors: | Fedorov, A.A, Fedorov, E.V, Samantha, D, Hillerich, B, Seidel, R.D, Almo, S.C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | CRYSTAL STRUCTURE OF HUMAN PD-L1
To Be Published
|
|
2ZAY
 
 | | Crystal structure of response regulator from Desulfuromonas acetoxidans | | Descriptor: | Response regulator receiver protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-12 | | Release date: | 2007-10-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of response regulator from Desulfuromonas acetoxidans.
To be Published
|
|
3V5F
 
 | | Crystal structure of the mutant E234A of Galacturonate Dehydratase from GEOBACILLUS SP. complexed with Mg | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Groninger-Poe, F, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-12-16 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the mutant E234A of Galacturonate Dehydratase from GEOBACILLUS SP. complexed with Mg
To be Published
|
|
3VE9
 
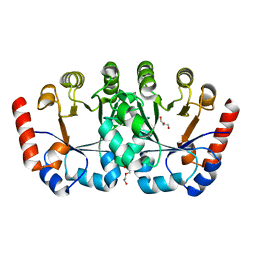 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Metallosphaera sedula | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Orotidine-5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-01-07 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Metallosphaera sedula
To be Published
|
|
3VE7
 
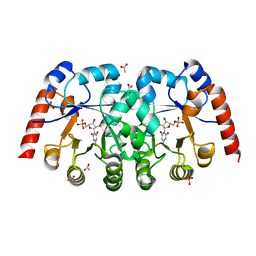 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Metallosphaera sedula complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, ACETIC ACID, Orotidine-5'-phosphate decarboxylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-01-07 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Metallosphaera sedula complexed with inhibitor BMP
To be Published
|
|
3VCC
 
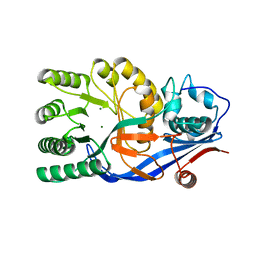 | | CRYSTAL STRUCTURE OF D-Galacturonate Dehydratase from GEOBACILLUS SP. complexed with Mg | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Groninger-Poe, F, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-01-03 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | CRYSTAL STRUCTURE OF D-Galacturonate Dehydratase from GEOBACILLUS SP. complexed with Mg
To be Published
|
|
1N10
 
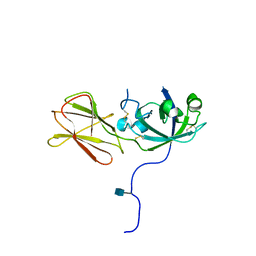 | | Crystal Structure of Phl p 1, a Major Timothy Grass Pollen Allergen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pollen allergen Phl p 1 | | Authors: | Fedorov, A.A, Ball, T, Leistler, B, Valenta, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-10-16 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray Crystal Structure of Phl p 1, a Major Timothy Grass Pollen Allergen
To be Published
|
|
3L8D
 
 | | Crystal structure of methyltransferase from Bacillus Thuringiensis | | Descriptor: | Methyltransferase, POTASSIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-30 | | Release date: | 2010-01-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of methyltransferase from Bacillus Thuringiensis
To be Published
|
|
3LKE
 
 | | Crystal structure of enoyl-CoA hydratase from Bacillus halodurans | | Descriptor: | Enoyl-CoA hydratase, GLYCEROL | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-27 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of enoyl-CoA hydratase from Bacillus halodurans
To be Published
|
|
3LSC
 
 | | Crystal structure of the mutant E241Q of atrazine chlorohydrolase TrzN from Arthrobacter aurescens TC1 complexed with zinc and atraton | | Descriptor: | N-ethyl-6-methoxy-N'-(1-methylethyl)-1,3,5-triazine-2,4-diamine, Triazine hydrolase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Seffernick, J, Wackett, L.P, Almo, S.C. | | Deposit date: | 2010-02-12 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of the mutant E241Q of atrazine chlorohydrolase
TrzN from Arthrobacter aurescens TC1 complexed with zinc and atraton
To be Published
|
|
3LS9
 
 | | Crystal structure of atrazine chlorohydrolase TrzN from Arthrobacter aurescens TC1 complexed with zinc | | Descriptor: | Triazine hydrolase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Seffernick, J, Wackett, L.P, Almo, S.C. | | Deposit date: | 2010-02-12 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of atrazine chlorohydrolase TrzN
from Arthrobacter aurescens TC1 complexed with zinc
To be Published
|
|
3LSB
 
 | | Crystal structure of the mutant E241Q of atrazine chlorohydrolase TrzN from Arthrobacter aurescens TC1 complexed with zinc and ametrin | | Descriptor: | N-ethyl-N'-(1-methylethyl)-6-(methylsulfanyl)-1,3,5-triazine-2,4-diamine, Triazine hydrolase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Seffernick, J, Wackett, L.P, Almo, S.C. | | Deposit date: | 2010-02-12 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Crystal structure of the mutant E241Q of atrazine chlorohydrolase TrzN
from Arthrobacter aurescens TC1 complexed with zinc and Ametryn
To be Published
|
|
3LY0
 
 | | Crystal structure of metallo peptidase from Rhodobacter sphaeroides liganded with phosphinate mimic of dipeptide L-Ala-D-Ala | | Descriptor: | (2R)-3-[(R)-[(1R)-1-aminoethyl](hydroxy)phosphoryl]-2-methylpropanoic acid, Dipeptidase AC. Metallo peptidase. MEROPS family M19, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Crystal structure of metallo peptidase from Rhodobacter sphaeroides
liganded with phosphinate mimic of dipeptide L-Ala-D-Ala
To be Published
|
|
1TZZ
 
 | | Crystal structure of the protein L1841, unknown member of enolase superfamily from Bradyrhizobium japonicum | | Descriptor: | Hypothetical protein L1841, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-12 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the protein L1841, unknown member of enolase superfamily from Bradyrhizobium japonicum
To be Published
|
|
1T62
 
 | |
1TEL
 
 | | Crystal structure of a RubisCO-like protein from Chlorobium tepidum | | Descriptor: | ribulose bisphosphate carboxylase, large subunit | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-05-25 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a RubisCO-like protein from Chlorobium tepidum
To be Published
|
|
1SV6
 
 | | Crystal structure of 2-hydroxypentadienoic acid hydratase from Escherichia Coli | | Descriptor: | 2-keto-4-pentenoate hydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sharp, A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-03-28 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 2-hydroxypentadienoic acid hydratase from Escherichia Coli
To be Published
|
|
1WHO
 
 | | ALLERGEN PHL P 2 | | Descriptor: | ALLERGEN PHL P 2 | | Authors: | Fedorov, A.A, Fedorov, E.V, Dolecek, C, Susani, M, Valenta, R, Almo, S.C. | | Deposit date: | 1997-04-04 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Phl P 2, a Major Timothy Grass (Phleum Pratense) Pollen Allergen
To be Published
|
|
1G5U
 
 | | LATEX PROFILIN HEVB8 | | Descriptor: | PROFILIN, SODIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Ganglberger, E, Breiteneder, H, Almo, S.C. | | Deposit date: | 2000-11-02 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Comparative Structural Analysis of Allergen Profilins HEVB8 and BETV2
To be Published
|
|
1WVI
 
 | |
