1O6Z
 
 | | 1.95 A resolution structure of (R207S,R292S) mutant of malate dehydrogenase from the halophilic archaeon Haloarcula marismortui (holo form) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Irimia, A, Ebel, C, Madern, D, Richard, S.B, Cosenza, L.W, Zaccai, G, Vellieux, F.M.D. | | Deposit date: | 2002-10-22 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Oligomeric States of Haloarcula Marismortui Malate Dehydrogenase are Modulated by Solvent Components as Shown by Crystallographic and Biochemical Studies
J.Mol.Biol., 326, 2003
|
|
2X0R
 
 | | R207S, R292S Mutant of Malate Dehydrogenase from the Halophilic Archeon Haloarcula marismortui (HoloForm) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Irimia, A, Ebel, C, Vellieux, F.M.D, Richard, S.B, Cosenza, L.W, Zaccai, G, Madern, D. | | Deposit date: | 2009-12-17 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.915 Å) | | Cite: | The Oligomeric States of Haloarcula Marismortui Malate Dehydrogenase are Modulated by Solvent Components as Shown by Crystallographic and Biochemical Studies
J.Mol.Biol., 326, 2003
|
|
3ZGH
 
 | | Crystal structure of the KRT10-binding region domain of the pneumococcal serine rich repeat protein PsrP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CELL WALL SURFACE ANCHOR FAMILY PROTEIN, ... | | Authors: | Schulte, T, Loefling, J, Mikaelsson, C, Kikhney, A, Hentrich, K, Diamante, A, Ebel, C, Normark, S, Svergun, D, Henriques-Normark, B, Achour, A. | | Deposit date: | 2012-12-17 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Basic Keratin 10-Binding Domain of the Virulence-Associated Pneumococcal Serine-Rich Protein Psrp Adopts a Novel Mscramm Fold.
Open Biol., 4, 2014
|
|
3ZGI
 
 | | Crystal structure of the KRT10-binding region domain of the pneumococcal serine rich repeat protein PsrP | | Descriptor: | 1,2-ETHANEDIOL, CELL WALL SURFACE ANCHOR FAMILY PROTEIN, SULFATE ION | | Authors: | Schulte, T, Loefling, J, Mikaelsson, C, Kikhney, A, Hentrich, K, Diamante, A, Ebel, C, Normark, S, Svergun, D, Henriques-Normark, B, Achour, A. | | Deposit date: | 2012-12-17 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Basic Keratin 10-Binding Domain of the Virulence-Associated Pneumococcal Serine-Rich Protein Psrp Adopts a Novel Mscramm Fold.
Open Biol., 4, 2014
|
|
3C22
 
 | |
5JUI
 
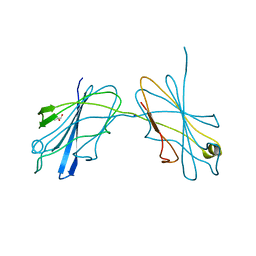 | |
4F7O
 
 | | Crystal structure of CSN5 | | Descriptor: | COP9 signalosome complex subunit 5, THIOCYANATE ION, ZINC ION | | Authors: | Echalier, A, Birol, M, Hoh, F, Dumas, C. | | Deposit date: | 2012-05-16 | | Release date: | 2013-01-23 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the regulation of the human COP9 signalosome catalytic subunit, CSN5/Jab1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2CF4
 
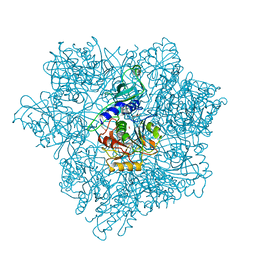 | | Pyrococcus horikoshii TET1 peptidase can assemble into a tetrahedron or a large octahedral shell | | Descriptor: | COBALT (II) ION, PROTEIN PH0519 | | Authors: | Vellieux, F.M.D, Schoehn, G, Dura, M.A, Roussel, A, Franzetti, B. | | Deposit date: | 2006-02-15 | | Release date: | 2006-09-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | An Archaeal Peptidase Assembles Into Two Different Quaternary Structures: A Tetrahedron and a Giant Octahedron.
J.Biol.Chem., 281, 2006
|
|
2K8J
 
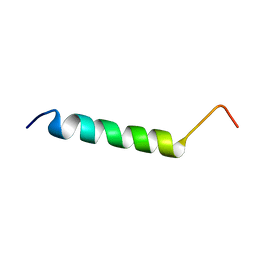 | | Solution structure of HCV p7 tm2 | | Descriptor: | p7tm2 | | Authors: | Montserret, R, Penin, F. | | Deposit date: | 2008-09-12 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure and ion channel activity of the p7 protein from hepatitis C virus.
J.Biol.Chem., 285, 2010
|
|
4DCS
 
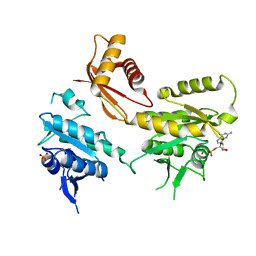 | | Crystal Structure of B. subtilis EngA in complex with sulfate ion and GDP | | Descriptor: | GTP-BINDING PROTEIN ENGA, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Reiser, J.-B, Housset, D, Foucher, A.-E, Jault, J.-M. | | Deposit date: | 2012-01-18 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Potassium Acts as a GTPase-Activating Element on Each Nucleotide-Binding Domain of the Essential Bacillus subtilis EngA.
Plos One, 7, 2012
|
|
4DCV
 
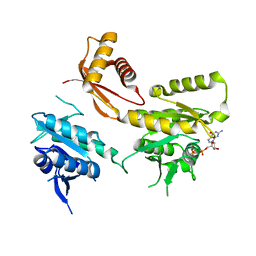 | | Crystal Structure of B. subtilis EngA in complex with GMPPCP | | Descriptor: | GTP-BINDING PROTEIN ENGA, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Reiser, J.-B, Housset, D, Foucher, A.-E, Jault, J.-M. | | Deposit date: | 2012-01-18 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Potassium Acts as a GTPase-Activating Element on Each Nucleotide-Binding Domain of the Essential Bacillus subtilis EngA.
Plos One, 7, 2012
|
|
2XR6
 
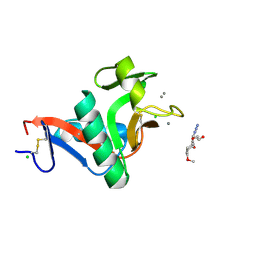 | | Crystal structure of the complex of the carbohydrate recognition domain of human DC-SIGN with pseudo trimannoside mimic. | | Descriptor: | 2-AZIDOETHANOL, CALCIUM ION, CD209 ANTIGEN, ... | | Authors: | Thepaut, M, Suitkeviciute, I, Sattin, S, Reina, J, Bernardi, A, Fieschi, F. | | Deposit date: | 2010-09-10 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Unique Dc-Sign Clustering Activity of a Small Glycomimetic: A Lesson for Ligand Design.
Acs Chem.Biol., 9, 2014
|
|
4DCT
 
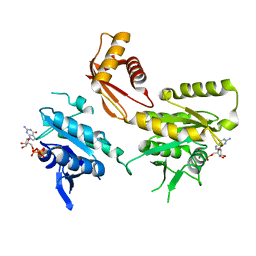 | | Crystal Structure of B. subtilis EngA in complex with half-occupacy GDP | | Descriptor: | GTP-BINDING PROTEIN ENGA, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Reiser, J.-B, Housset, D, Foucher, A.-E, Jault, J.-M. | | Deposit date: | 2012-01-18 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potassium Acts as a GTPase-Activating Element on Each Nucleotide-Binding Domain of the Essential Bacillus subtilis EngA.
Plos One, 7, 2012
|
|
4DCU
 
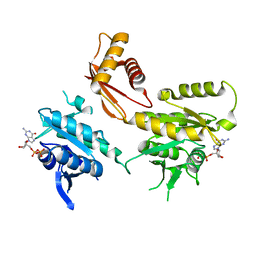 | | Crystal Structure of B. subtilis EngA in complex with GDP | | Descriptor: | GTP-BINDING PROTEIN ENGA, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Reiser, J.-B, Housset, D, Foucher, A.-E, Jault, J.-M. | | Deposit date: | 2012-01-18 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potassium Acts as a GTPase-Activating Element on Each Nucleotide-Binding Domain of the Essential Bacillus subtilis EngA.
Plos One, 7, 2012
|
|
4JCO
 
 | |
2J5R
 
 | | 2.25 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui after second radiation burn (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-19 | | Release date: | 2006-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
2J5Q
 
 | | 2.15 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui after first radiation burn (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-19 | | Release date: | 2006-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
2J5K
 
 | | 2.0 A resolution structure of the wild type malate dehydrogenase from Haloarcula marismortui (radiation damage series) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE | | Authors: | Fioravanti, E, Vellieux, F.M.D, Amara, P, Madern, D, Weik, M. | | Deposit date: | 2006-09-18 | | Release date: | 2006-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific Radiation Damage to Acidic Residues and its Relation to Their Chemical and Structural Environment.
J.Synchrotron Radiat., 14, 2007
|
|
2WZN
 
 | | 3d structure of TET3 from Pyrococcus horikoshii | | Descriptor: | 354AA LONG HYPOTHETICAL OPERON PROTEIN FRV, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rosenbaum, E, Dura, M.A, Vellieux, F.M, Franzetti, B. | | Deposit date: | 2009-12-01 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural and Biochemical Characterizations of a Novel Tet Peptidase Complex from Pyrococcus Horikoshii Reveal an Integrated Peptide Degradation System in Hyperthermophilic Archaea.
Mol.Microbiol., 72, 2009
|
|
2WYR
 
 | |
2X0I
 
 | | 2.9 A RESOLUTION STRUCTURE OF MALATE DEHYDROGENASE FROM ARCHAEOGLOBUS FULGIDUS IN COMPLEX WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MALATE DEHYDROGENASE, SODIUM ION, ... | | Authors: | Irimia, A, Madern, D, Zaccai, G, Vellieux, F.M.D, Karshikoff, A, Tibbelin, G, Ladenstein, R, Lien, T, Birkeland, N.-K. | | Deposit date: | 2009-12-14 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | The 2.9A Resolution Crystal Structure of Malate Dehydrogenase from Archaeoglobus Fulgidus: Mechanisms of Oligomerisation and Thermal Stabilisation.
J.Mol.Biol., 335, 2004
|
|
2X0J
 
 | | 2.8 A RESOLUTION STRUCTURE OF MALATE DEHYDROGENASE FROM ARCHAEOGLOBUS FULGIDUS IN COMPLEX WITH ETHENO-NAD | | Descriptor: | ETHENO-NAD, MALATE DEHYDROGENASE, SULFATE ION | | Authors: | Irimia, A, Madern, D, Zaccai, G, Vellieux, F.M, Karshikoff, A, Tibbelin, G, Ladenstein, R, Lien, T, Birkeland, N.K. | | Deposit date: | 2009-12-14 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.786 Å) | | Cite: | The 2.9A Resolution Crystal Structure of Malate Dehydrogenase from Archaeoglobus Fulgidus: Mechanisms of Oligomerisation and Thermal Stabilisation.
J.Mol.Biol., 335, 2004
|
|
4BGV
 
 | |
4BGU
 
 | | 1.50 A resolution structure of the malate dehydrogenase from Haloferax volcanii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Talon, R, Madern, D, Girard, E. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.487 Å) | | Cite: | Insight Into Structural Evolution of Extremophilic Proteins
To be Published
|
|
6GHV
 
 | | Structure of a DC-SIGN CRD in complex with high affinity glycomimetic. | | Descriptor: | CALCIUM ION, CD209 antigen, CHLORIDE ION, ... | | Authors: | Thepaut, M, Achilli, S, Medve, L, Bernardi, A, Fieschi, F. | | Deposit date: | 2018-05-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhancing Potency and Selectivity of a DC-SIGN Glycomimetic Ligand by Fragment-Based Design: Structural Basis.
Chemistry, 25, 2019
|
|
