6LUF
 
 | |
6LUA
 
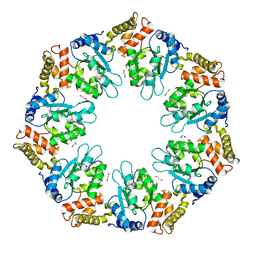 | |
5W1O
 
 | | Crystal Structure of HPV16 L1 Pentamer Bound to Heparin Oligosaccharides | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, Major capsid protein L1 | | Authors: | Dasgupta, J, Chen, X.S. | | Deposit date: | 2017-06-04 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of oligosaccharide receptor recognition by human papillomavirus.
J. Biol. Chem., 286, 2011
|
|
1NQP
 
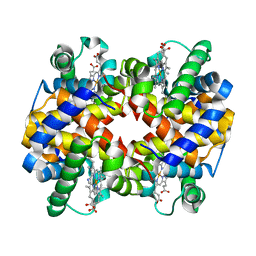 | | Crystal structure of Human hemoglobin E at 1.73 A resolution | | Descriptor: | CYANIDE ION, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Dasgupta, J, Sen, U, Choudhury, D, Dutta, P, Basu, S, Chakrabarti, A, Chakrabarty, A, Dattagupta, J.K. | | Deposit date: | 2003-01-22 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystallization and preliminary X-ray structural Studies of Hemoglobin A2 and Hemoglobin E, isolated from the blood samples of Beta-thalassemic patients
Biochem.Biophys.Res.Commun., 303, 2004
|
|
8X1A
 
 | | Crystal structure of periplasmic G6P binding protein VcA0625 | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, Iron(III) ABC transporter, periplasmic iron-compound-binding protein | | Authors: | Dasgupta, J, Saha, I. | | Deposit date: | 2023-11-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structural insights in to the atypical type-I ABC Glucose-6-phosphate importer VCA0625-27 of Vibrio cholerae.
Biochem.Biophys.Res.Commun., 716, 2024
|
|
7YRT
 
 | |
8H5V
 
 | | Crystal structure of the FleQ domain of Vibrio cholerae FlrA | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, flagellar regulatory protein A | | Authors: | Dasgupta, J, Chakraborty, S. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The N-terminal FleQ domain of the Vibrio cholerae flagellar master regulator FlrA plays pivotal structural roles in stabilizing its active state.
Febs Lett., 597, 2023
|
|
5W1X
 
 | | Crystal Structure of Humanpapillomavirus18 (HPV18) Capsid L1 Pentamers Bound to Heparin Oligosaccharides | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, ... | | Authors: | Chen, X.S, Dasgupta, J. | | Deposit date: | 2017-06-05 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.374 Å) | | Cite: | Structural basis of oligosaccharide receptor recognition by human papillomavirus.
J. Biol. Chem., 286, 2011
|
|
2ET2
 
 | | Crystal structure of an Asn to Ala mutant of Winged Bean Chymotrypsin Inhibitor protein | | Descriptor: | Chymotrypsin inhibitor 3, NICKEL (II) ION, SULFATE ION | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
2ESU
 
 | | Crystal structure of Asn to Gln mutant of Winged Bean Chymotrypsin Inhibitor protein | | Descriptor: | Chymotrypsin inhibitor 3, NICKEL (II) ION, SULFATE ION | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
2BEA
 
 | | Crystal structure of Asn14 to Gly mutant of WCI | | Descriptor: | Chymotrypsin inhibitor 3 | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-24 | | Release date: | 2006-06-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
2BEB
 
 | | X-ray structure of Asn to Thr mutant of Winged Bean Chymotrypsin inhibitor | | Descriptor: | Chymotrypsin inhibitor 3 | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-24 | | Release date: | 2006-06-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
4LX8
 
 | |
3TO5
 
 | |
2LMA
 
 | | Solution structure of CD4+ T cell derived peptide Thp5 | | Descriptor: | Thp5 peptide | | Authors: | Pandey, N.K, Khan, M.M, Chatterjee, S, Dwivedi, V.P, Tousif, S, Das, J, Van Kaer, L. | | Deposit date: | 2011-11-29 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | CD4+ T cell derived novel peptide Thp5 induces IL-4 production in CD4+ T cells to direct T helper 2 cell differentiation
J.Biol.Chem., 2011
|
|
4FKD
 
 | | Identification of the Activator Binding Residues in the Second Cysteine-Rich Regulatory Domain of Protein Kinase C Theta | | Descriptor: | Protein kinase C theta type, ZINC ION | | Authors: | Rahman, G.M, Shanker, S, Lewin, N.E, Prasad, B.V.V, Blumberg, P.M, Das, J. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.633 Å) | | Cite: | Identification of the Activator Binding Residues in the Second Cysteine-Rich Regulatory Domain of Protein Kinase C Theta.
Biochem.J., 451, 2013
|
|
7W61
 
 | | Crystal structure of farnesol dehydrogenase from Helicoverpa armigera | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Farnesol dehydrogenase, ... | | Authors: | Kumar, R, Das, J, Mahto, J.K, Sharma, M, Kumar, P, Sharma, A.K. | | Deposit date: | 2021-12-01 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and molecular characterization of NADP + -farnesol dehydrogenase from cotton bollworm, Helicoverpaarmigera.
Insect Biochem.Mol.Biol., 147, 2022
|
|
2QYI
 
 | | Crystal structure of a binary complex between an engineered trypsin inhibitor and Bovine trypsin | | Descriptor: | CALCIUM ION, Cationic trypsin, Chymotrypsin inhibitor 3, ... | | Authors: | Khamrui, S, Dasgupta, J, Dattagupta, J.K, Sen, U. | | Deposit date: | 2007-08-15 | | Release date: | 2008-08-19 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a binary complex between an engineered trypsin inhibitor and Bovine trypsin
To be Published
|
|
4QHT
 
 | | Crystal structure of AAA+/ sigma 54 activator domain of the flagellar regulatory protein FlrC from Vibrio cholerae in ATP analog bound state | | Descriptor: | 1,2-ETHANEDIOL, Flagellar regulatory protein C, MAGNESIUM ION, ... | | Authors: | Dey, S, Biswas, M, Sen, U, Dasgupta, J. | | Deposit date: | 2014-05-29 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.559 Å) | | Cite: | Unique ATPase site architecture triggers cis-mediated synchronized ATP binding in heptameric AAA+-ATPase domain of flagellar regulatory protein FlrC
J.Biol.Chem., 290, 2015
|
|
5GGX
 
 | |
5GGY
 
 | |
4H60
 
 | |
4HNQ
 
 | |
4HNS
 
 | | Crystal structure of activated CheY3 of Vibrio cholerae | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, MAGNESIUM ION | | Authors: | Biswas, M, Sen, U, Dasgupta, J. | | Deposit date: | 2012-10-21 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Barrier of CheY3 and Inability of CheY4 to Bind FliM Control the Flagellar Motor Action in Vibrio cholerae.
Plos One, 8, 2013
|
|
4HNR
 
 | |
