4NSS
 
 | | A structural and functional investigation of a novel protein from Mycobacterium smegmatis implicated in mycobacterial macrophage survivability | | Descriptor: | GLYCEROL, Mycobacterial protein | | Authors: | Shahine, A.E, Littler, D, Brammananath, R, Chan, P.Y, Crellin, P.K, Coppel, R.L, Rossjohn, J, Beddoe, T. | | Deposit date: | 2013-11-28 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural and functional investigation of a novel protein from Mycobacterium smegmatis implicated in mycobacterial macrophage survivability.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2WZM
 
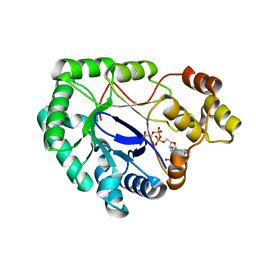 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-11-30 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
2WZT
 
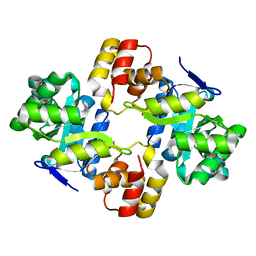 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-12-03 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
3AJA
 
 | |
7N3V
 
 | | Crystal structure of Mycobacterium smegmatis LmcA | | Descriptor: | GLYCEROL, LmcA, SULFATE ION | | Authors: | Patel, O, Lucet, I, Panjikar, S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the putative cell-wall lipoglycan biosynthesis protein LmcA from Mycobacterium smegmatis.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
2GRV
 
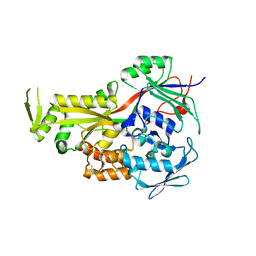 | | Crystal Structure of LpqW | | Descriptor: | LpqW | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hijacking of a Substrate-binding Protein Scaffold for use in Mycobacterial Cell Wall Biosynthesis
J.Mol.Biol., 359, 2006
|
|
7SHW
 
 | |
6UW1
 
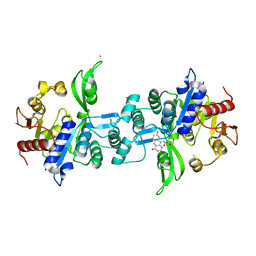 | | The crystal structure of FbiA from Mycobacterium Smegmatis, Fo bound form | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, CALCIUM ION, Phosphoenolpyruvate transferase | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6UVX
 
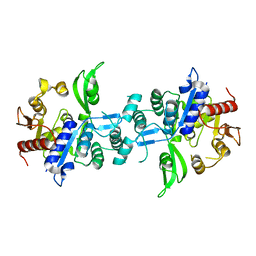 | | The crystal structure of FbiA from Mycobacterium Smegmatis, Apo state | | Descriptor: | CALCIUM ION, Phosphoenolpyruvate transferase | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6UW5
 
 | | The crystal structure of FbiA from Mycobacterium smegmatis, GDP and Fo bound form | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6UW7
 
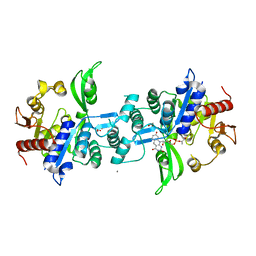 | | The crystal structure of FbiA from Mycobacterium smegmatis, Dehydro-F420-0 bound form | | Descriptor: | 2-[oxidanyl-[(2~{R},3~{S},4~{S})-2,3,4-tris(oxidanyl)-5-[2,4,8-tris(oxidanylidene)-1,9-dihydropyrimido[4,5-b]quinolin-10-yl]pentoxy]phosphoryl]oxyprop-2-enoic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Izore, T, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6UW3
 
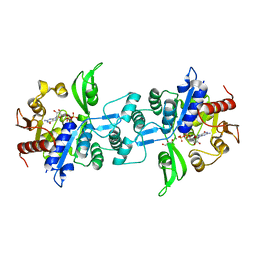 | | The crystal structure of FbiA from Mycobacterium Smegmatis, GDP Bound form | | Descriptor: | CALCIUM ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
5N29
 
 | |
3CKQ
 
 | | Crystal Structure of a Mycobacterial Protein | | Descriptor: | MANGANESE (II) ION, Putative uncharacterized protein, SULFATE ION, ... | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2008-03-16 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species.
J.Biol.Chem., 283, 2008
|
|
3CKV
 
 | | Crystal Structure of a Mycobacterial Protein | | Descriptor: | GLYCEROL, Putative uncharacterized protein, SULFATE ION, ... | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species.
J.Biol.Chem., 283, 2008
|
|
3CKO
 
 | | Crystal Structure of a Mycobacterial Protein | | Descriptor: | GLYCEROL, Putative uncharacterized protein, SULFATE ION, ... | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2008-03-16 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species.
J.Biol.Chem., 283, 2008
|
|
3CKN
 
 | | Crystal Structure of a Mycobacterial Protein | | Descriptor: | MANGANESE (II) ION, Putative uncharacterized protein, SULFATE ION, ... | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2008-03-16 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species.
J.Biol.Chem., 283, 2008
|
|
3CKJ
 
 | | Crystal Structure of a Mycobacterial Protein | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CITRIC ACID, PHOSPHATE ION, ... | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2008-03-15 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a UDP-glucose-specific glycosyltransferase from a Mycobacterium species.
J.Biol.Chem., 283, 2008
|
|
