1WVC
 
 | | alpha-D-glucose-1-phosphate cytidylyltransferase complexed with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Glucose-1-phosphate cytidylyltransferase, MAGNESIUM ION, ... | | Authors: | Koropatkin, N.M, Cleland, W.W, Holden, H.M. | | Deposit date: | 2004-12-14 | | Release date: | 2005-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinetic and structural analysis of alpha-D-Glucose-1-phosphate cytidylyltransferase from Salmonella typhi.
J.Biol.Chem., 280, 2005
|
|
1GG6
 
 | | CRYSTAL STRUCTURE OF GAMMA CHYMOTRYPSIN WITH N-ACETYL-PHENYLALANINE TRIFLUOROMETHYL KETONE BOUND AT THE ACTIVE SITE | | Descriptor: | 1,1,1-TRIFLUORO-3-ACETAMIDO-4-PHENYL BUTAN-2-ONE(N-ACETYL-L-PHENYLALANYL TRIFLUOROMETHYL KETONE), 1,2-ETHANEDIOL, GAMMA CHYMOTRYPSIN, ... | | Authors: | Neidhart, D, Wei, Y, Cassidy, C, Lin, J, Cleland, W.W, Frey, P.A. | | Deposit date: | 2000-07-31 | | Release date: | 2000-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Correlation of low-barrier hydrogen bonding and oxyanion binding in transition state analogue complexes of chymotrypsin.
Biochemistry, 40, 2001
|
|
1GGD
 
 | | CRYSTAL STRUCTURE OF GAMMA CHYMOTRYPSIN WITH N-ACETYL-LEUCIL-PHENYLALANINE ALDEHYDE BOUND AT THE ACTIVE SITE | | Descriptor: | 2-ACETYLAMINO-4-METHYL-PENTANOIC ACID (1-FORMYL-2-PHENYL-ETHYL)-AMIDE, GAMMA CHYMOTRYPSIN, SULFATE ION | | Authors: | Neidhart, D, Wei, Y, Cassidy, C, Lin, J, Cleland, W.W, Frey, P.A. | | Deposit date: | 2000-08-14 | | Release date: | 2000-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Correlation of low-barrier hydrogen bonding and oxyanion binding in transition state analogue complexes of chymotrypsin.
Biochemistry, 40, 2000
|
|
3S0M
 
 | | A Structural Element that Modulates Proton-Coupled Electron Transfer in Oxalate Decarboxylase | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saylor, B.T, Reinhardt, L.A, Lu, Z, Shukla, M.S, Cleland, W.W, Allen, K.N, Richards, N.G.J. | | Deposit date: | 2011-05-13 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A structural element that facilitates proton-coupled electron transfer in oxalate decarboxylase.
Biochemistry, 51, 2012
|
|
1P48
 
 | | REVERSE PROTONATION IS THE KEY TO GENERAL ACID-BASE CATALYSIS IN ENOLASE | | Descriptor: | Enolase 1, MAGNESIUM ION, PHOSPHOENOLPYRUVATE | | Authors: | Sims, P.A, Larsen, T.M, Poyner, R.R, Cleland, W.W, Reed, G.H. | | Deposit date: | 2003-04-21 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reverse protonation is the key to general acid-base catalysis in enolase
Biochemistry, 42, 2003
|
|
1P43
 
 | | REVERSE PROTONATION IS THE KEY TO GENERAL ACID-BASE CATALYSIS IN ENOLASE | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, Enolase 1, MAGNESIUM ION | | Authors: | Sims, P.A, Larsen, T.M, Poyner, R.R, Cleland, W.W, Reed, G.H. | | Deposit date: | 2003-04-21 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reverse protonation is the key to general acid-base catalysis in enolase
Biochemistry, 42, 2003
|
|
1R66
 
 | | Crystal Structure of DesIV (dTDP-glucose 4,6-dehydratase) from Streptomyces venezuelae with NAD and TYD bound | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TDP-glucose-4,6-dehydratase, ... | | Authors: | Allard, S.T.M, Cleland, W.W, Holden, H.M. | | Deposit date: | 2003-10-14 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | High Resolution X-ray Structure of dTDP-Glucose 4,6-Dehydratase from Streptomyces venezuelae
J.Biol.Chem., 279, 2004
|
|
1R6D
 
 | | Crystal Structure of DesIV double mutant (dTDP-glucose 4,6-dehydratase) from Streptomyces venezuelae with NAD and DAU bound | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TDP-glucose-4,6-dehydratase | | Authors: | Allard, S.T.M, Cleland, W.W, Holden, H.M. | | Deposit date: | 2003-10-14 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High Resolution X-ray Structure of dTDP-Glucose 4,6-Dehydratase from Streptomyces venezuelae
J.Biol.Chem., 279, 2004
|
|
4EA8
 
 | | X-ray crystal structure of PerB from Caulobacter crescentus in complex with coenzyme A and GDP-N-acetylperosamine at 1 Angstrom resolution | | Descriptor: | CHLORIDE ION, COENZYME A, GDP-N-acetylperosamine, ... | | Authors: | Thoden, J.B, Reinhardt, L.A, Cook, P.D, Menden, P, Cleland, W.W, Holden, H.M. | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Catalytic Mechanism of Perosamine N-Acetyltransferase Revealed by High-Resolution X-ray Crystallographic Studies and Kinetic Analyses.
Biochemistry, 51, 2012
|
|
4EAA
 
 | | X-ray crystal structure of the H141N mutant of perosamine N-acetyltransferase from Caulobacter crescentus in complex with CoA and GDP-perosamine | | Descriptor: | CHLORIDE ION, COENZYME A, GDP-perosamine, ... | | Authors: | Thoden, J.B, Reinhardt, L.A, Cook, P.D, Menden, P, Cleland, W.W, Holden, H.M. | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Catalytic Mechanism of Perosamine N-Acetyltransferase Revealed by High-Resolution X-ray Crystallographic Studies and Kinetic Analyses.
Biochemistry, 51, 2012
|
|
4EAB
 
 | | X-ray crystal structure of the H141A mutant of GDP-perosamine N-acetyl transferase from Caulobacter crescentus in complex with CoA and GDP-perosamine | | Descriptor: | CHLORIDE ION, COENZYME A, GDP-perosamine, ... | | Authors: | Thoden, J.B, Reinhardt, L.A, Cook, P.D, Menden, P, Cleland, W.W, Holden, H.M. | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Catalytic Mechanism of Perosamine N-Acetyltransferase Revealed by High-Resolution X-ray Crystallographic Studies and Kinetic Analyses.
Biochemistry, 51, 2012
|
|
4EA7
 
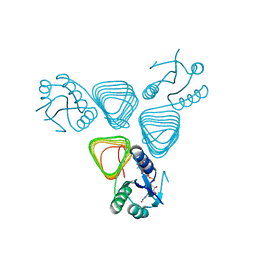 | | X-ray crystal structure of PerB from Caulobacter crescentus in complex with CoA and GDP-perosamine at 1.0 Angstrom resolution | | Descriptor: | CHLORIDE ION, COENZYME A, GDP-perosamine, ... | | Authors: | Thoden, J.B, Reinhardt, L.A, Cook, P.D, Menden, P, Cleland, W.W, Holden, H.M. | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Catalytic Mechanism of Perosamine N-Acetyltransferase Revealed by High-Resolution X-ray Crystallographic Studies and Kinetic Analyses.
Biochemistry, 51, 2012
|
|
4EA9
 
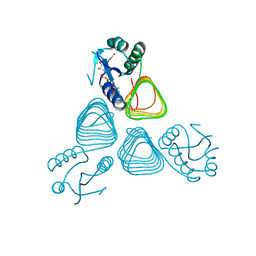 | | X-ray structure of GDP-perosamine N-acetyltransferase in complex with transition state analog at 0.9 Angstrom resolution | | Descriptor: | CHLORIDE ION, GDP-N-acetylperosamine-coenzyme A, Perosamine N-acetyltransferase | | Authors: | Thoden, J.B, Reinhardt, L.A, Cook, P.D, Menden, P, Cleland, W.W, Holden, H.M. | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Catalytic Mechanism of Perosamine N-Acetyltransferase Revealed by High-Resolution X-ray Crystallographic Studies and Kinetic Analyses.
Biochemistry, 51, 2012
|
|
3V8E
 
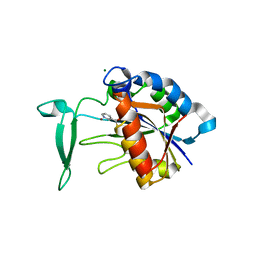 | | Crystal structure of the yeast nicotinamidase Pnc1p bound to the inhibitor nicotinaldehyde | | Descriptor: | MAGNESIUM ION, Nicotinamidase, ZINC ION | | Authors: | Hoadley, K.A, Smith, B.C, Denu, J.M, Keck, J.L. | | Deposit date: | 2011-12-22 | | Release date: | 2012-01-25 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural and Kinetic Isotope Effect Studies of Nicotinamidase (Pnc1) from Saccharomyces cerevisiae.
Biochemistry, 51, 2012
|
|
2QF7
 
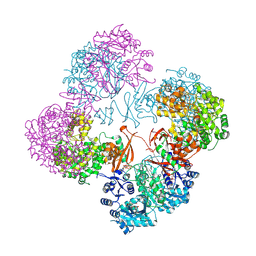 | | Crystal structure of a complete multifunctional pyruvate carboxylase from Rhizobium etli | | Descriptor: | CHLORIDE ION, COENZYME A, FORMIC ACID, ... | | Authors: | St Maurice, M, Surinya, K.H, Rayment, I. | | Deposit date: | 2007-06-27 | | Release date: | 2007-09-04 | | Last modified: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Domain architecture of pyruvate carboxylase, a biotin-dependent multifunctional enzyme
Science, 317, 2007
|
|
3TW7
 
 | | Structure of Rhizobium etli pyruvate carboxylase T882A crystallized without acetyl coenzyme-A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Pyruvate carboxylase protein, ... | | Authors: | St Maurice, M, Kumar, S, Lietzan, A.D. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-12 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Interaction between the biotin carboxyl carrier domain and the biotin carboxylase domain in pyruvate carboxylase from Rhizobium etli.
Biochemistry, 50, 2011
|
|
3TW6
 
 | | Structure of Rhizobium etli pyruvate carboxylase T882A with the allosteric activator, acetyl coenzyme-A | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | St Maurice, M, Kumar, S, Lietzan, A.D. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-19 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interaction between the biotin carboxyl carrier domain and the biotin carboxylase domain in pyruvate carboxylase from Rhizobium etli.
Biochemistry, 50, 2011
|
|
1FNJ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CHORISMATE MUTASE MUTANT C88S/R90K | | Descriptor: | PROTEIN (CHORISMATE MUTASE) | | Authors: | Kast, P, Grisostomi, C, Chen, I.A, Li, S, Krengel, U, Xue, Y, Hilvert, D. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A strategically positioned cation is crucial for efficient catalysis by chorismate mutase.
J.Biol.Chem., 275, 2000
|
|
1FNK
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CHORISMATE MUTASE MUTANT C88K/R90S | | Descriptor: | PROTEIN (CHORISMATE MUTASE) | | Authors: | Kast, P, Grisostomi, C, Chen, I.A, Li, S, Krengel, U, Xue, Y, Hilvert, D. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A strategically positioned cation is crucial for efficient catalysis by chorismate mutase.
J.Biol.Chem., 275, 2000
|
|
