7XLM
 
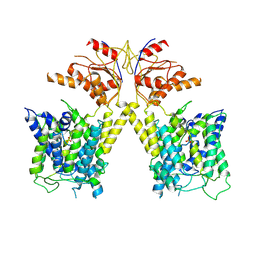 | |
7XUL
 
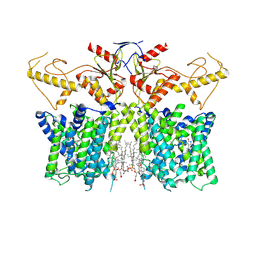 | | Human SLC26A3 in complex with Tenidap | | Descriptor: | 5-chloranyl-2-oxidanyl-3-thiophen-2-ylcarbonyl-indole-1-carboxamide, CHOLESTEROL HEMISUCCINATE, Chloride anion exchanger | | Authors: | Li, X.R, Chi, X.M, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure of human SLC26A3 in complex with Tenidap
To Be Published
|
|
7XUJ
 
 | | Human SLC26A3 in complex with UK5099 | | Descriptor: | (E)-2-cyano-3-(1-phenylindol-3-yl)prop-2-enoic acid, CHOLESTEROL HEMISUCCINATE, Chloride anion exchanger | | Authors: | Li, X.R, Chi, X.M, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | human SLC26A3 in complex with UK5099
To Be Published
|
|
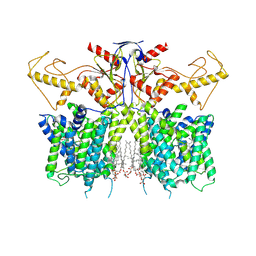
7XUH
 
 | | Down-regulated in adenoma in complex with TQR1122 | | Descriptor: | 2-[4,8-dimethyl-2-oxidanylidene-7-[[3-(trifluoromethyl)phenyl]methoxy]chromen-3-yl]ethanoic acid, CHOLESTEROL HEMISUCCINATE, Chloride anion exchanger | | Authors: | Li, X.R, Chi, X.M, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-05-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | structure of human SLC26A3 in complex with TQR1122
To Be Published
|
|
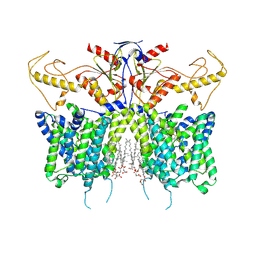
8IET
 
 | |
6M22
 
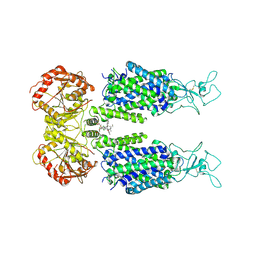 | | KCC3 bound with DIOA | | Descriptor: | 2-[[(2~{R})-2-butyl-6,7-bis(chloranyl)-2-cyclopentyl-1-oxidanylidene-3~{H}-inden-5-yl]oxy]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chi, X.M, Li, X.R, Chen, Y, Zhang, Y.Y, Su, Q, Zhou, Q. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-04 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of the full-length human KCC2 and KCC3 cation-chloride cotransporters.
Cell Res., 31, 2021
|
|
6M23
 
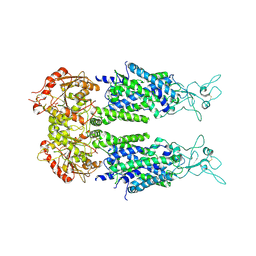 | | Overall structure of KCC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Chi, X.M, Li, X.R, Chen, Y, Zhang, Y.Y, Su, Q, Zhou, Q. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-04 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the full-length human KCC2 and KCC3 cation-chloride cotransporters.
Cell Res., 31, 2021
|
|
6M1Y
 
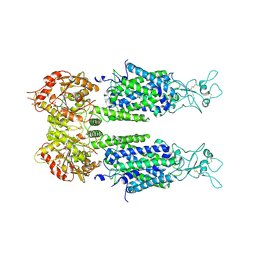 | | The overall structure of KCC3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Chi, X.M, Li, X.R, Chen, Y, Zhang, Y.Y, Su, Q, Zhou, Q. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the full-length human KCC2 and KCC3 cation-chloride cotransporters.
Cell Res., 31, 2021
|
|
7WWB
 
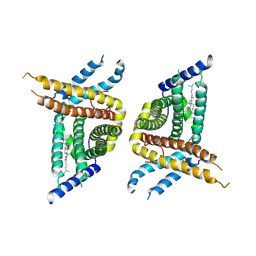 | | Choline transporter-like protein 1 | | Descriptor: | CHOLESTEROL, CHOLINE ION, Choline transporter-like protein 1 | | Authors: | Chi, X.M, Zhou, Q. | | Deposit date: | 2022-02-12 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Rational exploration of fold atlas for human solute carrier proteins.
Structure, 30, 2022
|
|
7CGN
 
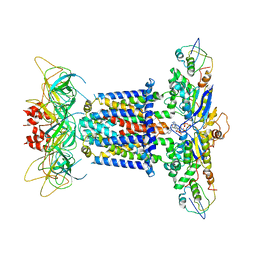 | | The overall structure of the MlaFEDB complex in ATP-bound EQtall conformation (Mutation of E170Q on MlaF) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | Authors: | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-07-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
7CGE
 
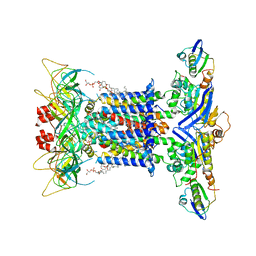 | | The overall structure of nucleotide free MlaFEDB complex | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | Authors: | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-07-01 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
7CH1
 
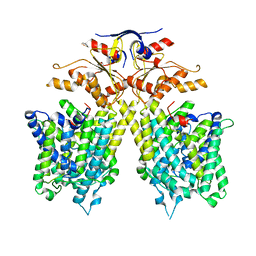 | | The overall structure of SLC26A9 | | Descriptor: | CHLORIDE ION, SODIUM ION, Solute carrier family 26 member 9 | | Authors: | Chi, X.M, Chen, Y, Li, X.R, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into the gating mechanism of human SLC26A9 mediated by its C-terminal sequence.
Cell Discov, 6, 2020
|
|
7CH0
 
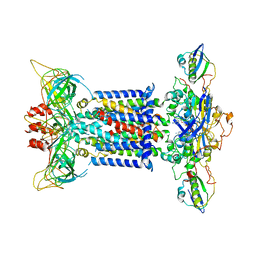 | | The overall structure of the MlaFEDB complex in ATP-bound EQclose conformation (Mutation of E170Q on MlaF) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, ... | | Authors: | Chi, X.M, Fan, Q.X, Zhang, Y.Y, Liang, K, Zhou, Q, Li, Y.Y. | | Deposit date: | 2020-07-03 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural mechanism of phospholipids translocation by MlaFEDB complex.
Cell Res., 30, 2020
|
|
6JGZ
 
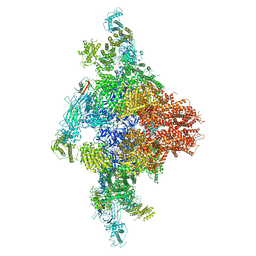 | | Structure of RyR2 (F/P/Ca2+ dataset) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, RyR2, ZINC ION | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-16 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JG3
 
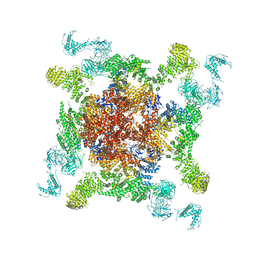 | | Cryo-EM structure of RyR2 (Ca2+ alone dataset) | | Descriptor: | Ryanodine receptor 2, ZINC ION | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-13 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JHN
 
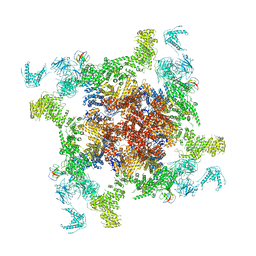 | | Structure of RyR2 (F/C/Ca2+ dataset) | | Descriptor: | CAFFEINE, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-18 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JH6
 
 | | Structure of RyR2 (F/A/Ca2+ dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Peptidyl-prolyl cis-trans isomerase FKBP1B, RyR2, ... | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-17 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8GTO
 
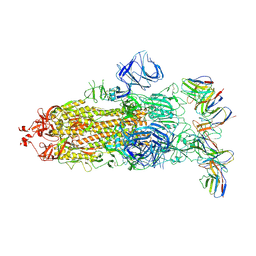 | | cryo-EM structure of Omicron BA.5 S protein in complex with XGv282 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
8GTP
 
 | | cryo-EM structure of Omicron BA.5 S protein in complex with XGv289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
8GTQ
 
 | | cryo-EM structure of Omicron BA.5 S protein in complex with S2L20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
7E21
 
 | | Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state with ATP-gamma-S | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7E1Z
 
 | | Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7E20
 
 | | Cryo EM structure of a K+-bound Na+,K+-ATPase in the E2 state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7DX5
 
 | | S protein of SARS-CoV-2 bound with PD of ACE2 in the conformation 2 (1 up RBD and 1 PD bound) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
7DX0
 
 | | Trypsin-digested S protein of SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-31 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
