2VGY
 
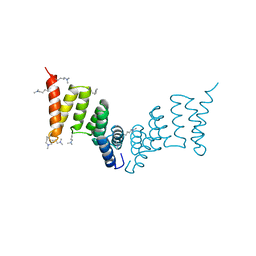 | | Crystal structure of the Yersinia enterocolitica Type III Secretion Translocator Chaperone SycD (alternative dimer) | | Descriptor: | CHAPERONE SYCD | | Authors: | Buttner, C.R, Sorg, I, Cornelis, G.R, Heinz, D.W, Niemann, H.H. | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Yersinia Enterocolitica Type III Secretion Chaperone Sycd
J.Mol.Biol., 375, 2008
|
|
2VGX
 
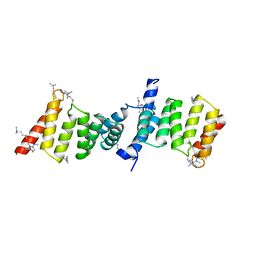 | | Structure of the Yersinia enterocolitica Type III Secretion Translocator Chaperone SycD | | Descriptor: | CHAPERONE SYCD | | Authors: | Buttner, C.R, Sorg, I, Cornelis, G.R, Heinz, D.W, Niemann, H.H. | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Yersinia Enterocolitica Type III Secretion Chaperone Sycd
J.Mol.Biol., 375, 2008
|
|
2BSI
 
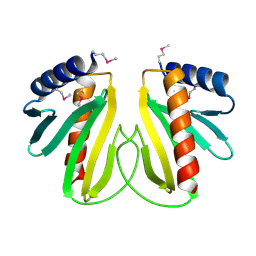 | |
2BSH
 
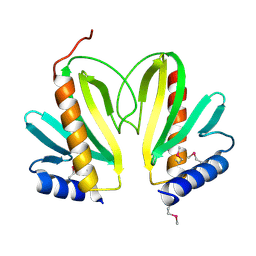 | |
2BSJ
 
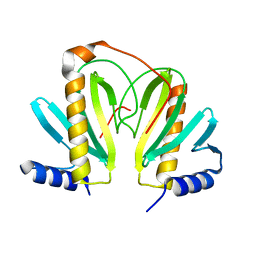 | | Native Crystal Structure of the Type III Secretion chaperone SycT from Yersinia enterocolitica | | Descriptor: | CHAPERONE PROTEIN SYCT, CHLORIDE ION | | Authors: | Buttner, C.R, Cornelis, G.R, Heinz, D.W, Niemann, H.H. | | Deposit date: | 2005-05-21 | | Release date: | 2005-09-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Yersinia Enterocolitica Type III Secretion Chaperone Syct.
Protein Sci., 14, 2005
|
|
3ZQO
 
 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 3) | | Descriptor: | POTASSIUM ION, TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZQP
 
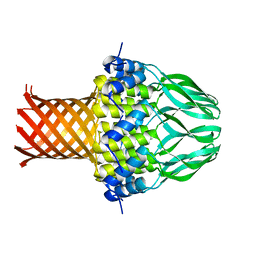 | | Crystal structure of the small terminase oligomerization domain from a SPP1-like bacteriophage | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZQQ
 
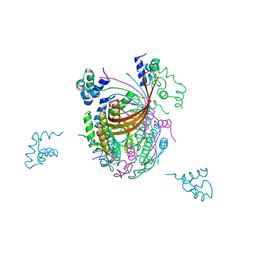 | | Crystal structure of the full-length small terminase from a SPP1-like bacteriophage | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZQN
 
 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 2) | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZQM
 
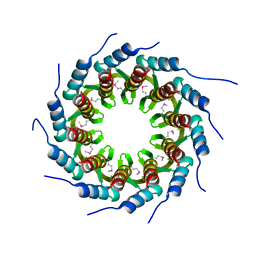 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 1) | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7QVY
 
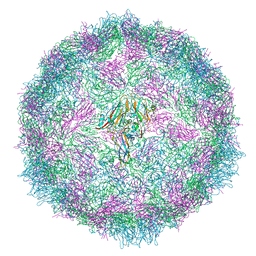 | | Cryo-EM structure of coxsackievirus A6 empty particle | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Buttner, C.R, Spurny, R, Fuzik, T, Plevka, P. | | Deposit date: | 2022-01-24 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Cryo-electron microscopy and image classification reveal the existence and structure of the coxsackievirus A6 virion.
Commun Biol, 5, 2022
|
|
7QW9
 
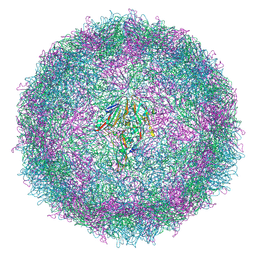 | | Cryo-EM structure of coxsackievirus A6 mature virion | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Buttner, C.R, Spurny, R, Fuzik, T, Plevka, P. | | Deposit date: | 2022-01-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Cryo-electron microscopy and image classification reveal the existence and structure of the coxsackievirus A6 virion.
Commun Biol, 5, 2022
|
|
7QVX
 
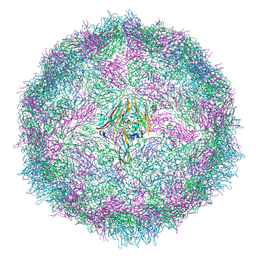 | | Cryo-EM structure of coxsackievirus A6 altered particle | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Buttner, C.R, Spurny, R, Fuzik, T, Plevka, P. | | Deposit date: | 2022-01-24 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-electron microscopy and image classification reveal the existence and structure of the coxsackievirus A6 virion.
Commun Biol, 5, 2022
|
|
8RBS
 
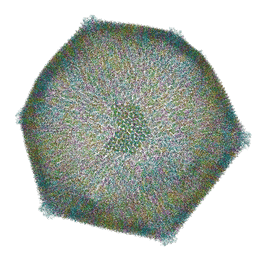 | | Emiliania huxleyi virus 201 (EhV-201) asymmetrical unit of capsid proteins predicted by AlphaFold2 fitted into the cryo-EM density of EhV-201 virion composite map. | | Descriptor: | Major capsid protein, Penton protein | | Authors: | Homola, M, Buttner, C.R, Fuzik, T, Novacek, J, Chaillet, M, Forster, F, Plevka, P. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structure and replication cycle of a virus infecting climate-modulating alga Emiliania huxleyi.
Sci Adv, 10, 2024
|
|
8RBT
 
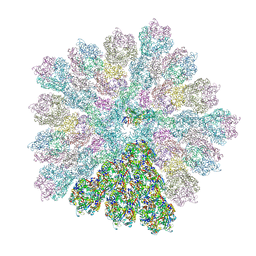 | | Emiliania huxleyi virus 201 (EhV-201) capsid proteins predicted by AlphaFold2 fitted into a cryo-EM density map of the EhV-201 virion capsid. | | Descriptor: | Major capsid protein, Penton protein | | Authors: | Homola, M, Buttner, C.R, Fuzik, T, Novacek, J, Chaillet, M, Forster, F, Plevka, P. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure and replication cycle of a virus infecting climate-modulating alga Emiliania huxleyi.
Sci Adv, 10, 2024
|
|
5YVQ
 
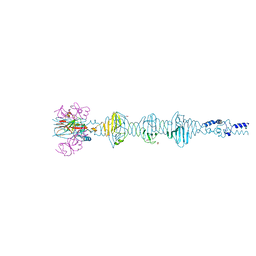 | | Complex of Mu phage tail fiber and its chaperone | | Descriptor: | GLYCEROL, Tail fiber assembly protein U, Tail fiber protein S | | Authors: | Takeda, S, Sakai, K, Iwazaki, T, Yamashita, E, Nakagawa, A. | | Deposit date: | 2017-11-27 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Phage tail fibre assembly proteins employ a modular structure to drive the correct folding of diverse fibres.
Nat Microbiol, 4, 2019
|
|
