6PQG
 
 | |
6PQF
 
 | |
5UJR
 
 | | NMR Solution Structure of the Two-component Bacteriocin CbnXY | | Descriptor: | Bacteriocin | | Authors: | Acedo, J.Z, Towle, K.M, Lohans, C.T, McKay, R.T, Miskolzie, M, Doerksen, T, Vederas, J.C, Martin-Visscher, L.A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification and three-dimensional structure of carnobacteriocin XY, a class IIb bacteriocin produced by Carnobacteria.
FEBS Lett., 591, 2017
|
|
5UJQ
 
 | | NMR Solution Structure of the Two-component Bacteriocin CbnXY | | Descriptor: | Bacteriocin | | Authors: | Acedo, J.Z, Towle, K.M, Lohans, C.T, McKay, R.T, Miskolzie, M, Doerksen, T, Vederas, J.C, Martin-Visscher, L.A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification and three-dimensional structure of carnobacteriocin XY, a class IIb bacteriocin produced by Carnobacteria.
FEBS Lett., 591, 2017
|
|
2N8O
 
 | | NMR Solution Structure of Aureocin A53 | | Descriptor: | Bacteriocin aureocin A53 | | Authors: | Acedo, J.Z, van Belkum, M.J, Lohans, C.T, Towle, K.M, Miskolzie, M, Vederas, J.C. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structures of Lacticin Q and Aureocin A53 Reveal a Structural Motif Conserved among Leaderless Bacteriocins with Broad-Spectrum Activity.
Biochemistry, 55, 2016
|
|
2N8P
 
 | | Solution Structure of Lacticin Q | | Descriptor: | Lacticin Q | | Authors: | Acedo, J.Z, van Belkum, M.J, Lohans, C.T, Towle, K.M, Miskolzie, M, Vederas, J.C. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structures of Lacticin Q and Aureocin A53 Reveal a Structural Motif Conserved among Leaderless Bacteriocins with Broad-Spectrum Activity.
Biochemistry, 55, 2016
|
|
5UZL
 
 | | Brassica napus DGAT1 exosite | | Descriptor: | O-acyltransferase | | Authors: | Acedo, J.Z, Vederas, J.C. | | Deposit date: | 2017-02-26 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Diacylglycerol Acyltransferase 1 Is Regulated by Its N-Terminal Domain in Response to Allosteric Effectors.
Plant Physiol., 175, 2017
|
|
2MWR
 
 | | Solution Structure of Acidocin B, a Circular Bacteriocin from Lactobacillus acidophilus M46 | | Descriptor: | Acidocin B | | Authors: | Vederas, J.C, Acedo, J.Z, van Belkum, M.J, Lohans, C.T. | | Deposit date: | 2014-11-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Acidocin B, a Circular Bacteriocin Produced by Lactobacillus acidophilus M46.
Appl.Environ.Microbiol., 81, 2015
|
|
6VLJ
 
 | |
5KGY
 
 | | Phenol-soluble modulin Alpha 3 | | Descriptor: | Phenol-soluble modulin alpha 3 peptide | | Authors: | Towle, K.M, Lohans, C.T, Acedo, J.Z, Van Belkum, M.J, Miskolzie, M, Vederas, J.C. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Phenol-Soluble Modulins alpha 1, alpha 3, and beta 2, Virulence Factors from Staphylococcus aureus.
Biochemistry, 55, 2016
|
|
5KHB
 
 | | Structure of Phenol-soluble modulin Alpha1 | | Descriptor: | PSM Alpha1 | | Authors: | Towle, K.M, Lohans, C.T, Acedo, J.Z, Miskolzie, M, van Belkum, M.J, Vederas, J.C. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Phenol-Soluble Modulins alpha 1, alpha 3, and beta 2, Virulence Factors from Staphylococcus aureus.
Biochemistry, 55, 2016
|
|
5KGZ
 
 | | Phenol-soluble modulin Beta2 | | Descriptor: | Modulin Beta2 | | Authors: | Towle, K.M, Lohans, C.T, Acedo, J.Z, Van Belkum, M.J, Miskolzie, M, Vederas, J.C. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Phenol-Soluble Modulins alpha 1, alpha 3, and beta 2, Virulence Factors from Staphylococcus aureus.
Biochemistry, 55, 2016
|
|
2N5W
 
 | | The NMR solution structure of octyl-tridecaptin A1 in DPC micelles | | Descriptor: | Octyl-tridecaptin A1 | | Authors: | Cochrane, S.A, Findlay, B, Bakhtiary, A, Acedo, J.Z, Rodriguez-Lopez, E.M, Vederas, J.C. | | Deposit date: | 2015-08-01 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Antimicrobial lipopeptide tridecaptin A1 selectively binds to Gram-negative lipid II.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7JVF
 
 | |
6VGT
 
 | |
6VHJ
 
 | |
6VE9
 
 | |
6VJQ
 
 | |
7JU9
 
 | |
6UAK
 
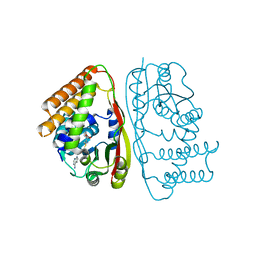 | | LahSb - C-terminal methyltransferase involved in RiPP biosynthesis | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM dependent methyltransferase LahSB | | Authors: | Nair, S.K, Estrada, P. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Characterization of a Dehydratase and Methyltransferase in the Biosynthesis of Ribosomally Synthesized and Post-translationally Modified Peptides in Lachnospiraceae.
Chembiochem, 21, 2020
|
|
2N5Y
 
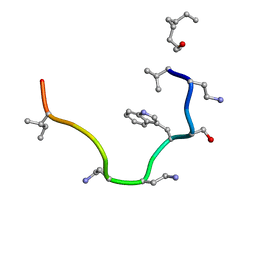 | | Solution NMR structure of octyl-tridecaptin A1 in DPC micelles containing Gram-negative lipid II | | Descriptor: | Octyl-tridecaptin A1 | | Authors: | Cochrane, S.A, Findlay, B, Bakhtiary, A, Rodriguez-Lopez, E.M, Vederas, J.C. | | Deposit date: | 2015-08-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Antimicrobial lipopeptide tridecaptin A1 selectively binds to Gram-negative lipid II.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
