4RCR
 
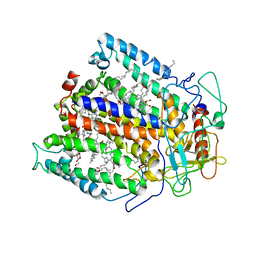 | | STRUCTURE OF THE REACTION CENTER FROM RHODOBACTER SPHAEROIDES R-26 AND 2.4.1: PROTEIN-COFACTOR (BACTERIOCHLOROPHYLL, BACTERIOPHEOPHYTIN, AND CAROTENOID) INTERACTIONS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Komiya, H, Yeates, T.O, Chirino, A.J, Rees, D.C, Allen, J.P, Feher, G. | | Deposit date: | 1991-09-09 | | Release date: | 1993-10-31 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the reaction center from Rhodobacter sphaeroides R-26 and 2.4.1: protein-cofactor (bacteriochlorophyll, bacteriopheophytin, and carotenoid) interactions.
Proc.Natl.Acad.Sci.USA, 85, 1988
|
|
5D6V
 
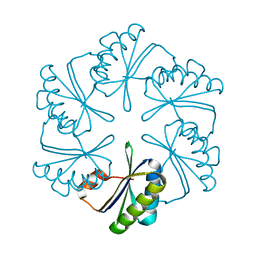 | |
1CYJ
 
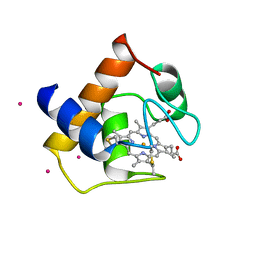 | | CYTOCHROME C6 | | Descriptor: | CADMIUM ION, CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 1995-05-09 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of chloroplast cytochrome c6 at 1.9 A resolution: evidence for functional oligomerization.
J.Mol.Biol., 250, 1995
|
|
3BN4
 
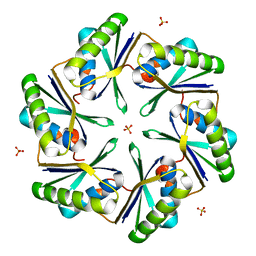 | | Carboxysome Subunit, CcmK1 | | Descriptor: | Carbon dioxide-concentrating mechanism protein ccmK homolog 1, SULFATE ION | | Authors: | Tanaka, S, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2007-12-13 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic-level models of the bacterial carboxysome shell.
Science, 319, 2008
|
|
2G13
 
 | | CsoS1A with sulfate ion | | Descriptor: | Major carboxysome shell protein 1A, SULFATE ION | | Authors: | Tsai, Y, Sawaya, M.R, Cannon, G.C, Williams, E.B, Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 2006-02-13 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural Analysis of CsoS1A and the Protein Shell of the Halothiobacillus neapolitanus Carboxysome.
Plos Biol., 5, 2007
|
|
4D9J
 
 | |
5UI2
 
 | | CRYSTAL STRUCTURE OF ORANGE CAROTENOID PROTEIN | | Descriptor: | (3'R)-3'-hydroxy-beta,beta-caroten-4-one, CHLORIDE ION, Orange carotenoid-binding protein, ... | | Authors: | KERFELD, C.A, SAWAYA, M.R, VISHNU, B, KROGMANN, D, YEATES, T.O. | | Deposit date: | 2017-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a cyanobacterial water-soluble carotenoid binding protein.
Structure, 11, 2003
|
|
2Z9H
 
 | | Ethanolamine utilization protein, EutN | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CHLORIDE ION, Ethanolamine utilization protein eutN | | Authors: | Tanaka, S, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The crystal structure of ethanolamine utilization protein EutN from E. coli
To be Published
|
|
6C9I
 
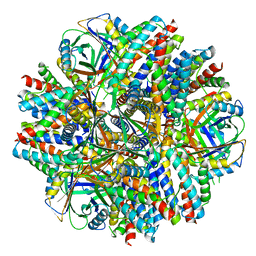 | | Single-Particle reconstruction of DARP14 - A designed protein scaffold displaying ~17kDa DARPin proteins - Scaffold | | Descriptor: | DARP14 - Subunit A with DARPin, DARP14 - Subunit B | | Authors: | Gonen, S, Liu, Y, Yeates, T.O, Gonen, T. | | Deposit date: | 2018-01-26 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Near-atomic cryo-EM imaging of a small protein displayed on a designed scaffolding system.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2QW7
 
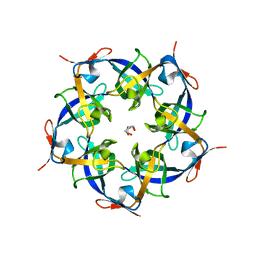 | | Carboxysome Subunit, CcmL | | Descriptor: | Carbon dioxide concentrating mechanism protein ccmL, GLYCEROL | | Authors: | Tanaka, S, Sawaya, M.R, Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 2007-08-09 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic-level models of the bacterial carboxysome shell.
Science, 319, 2008
|
|
6XPI
 
 | | CutR flat hexamer, form 1 | | Descriptor: | Ethanolamine utilization protein EutS | | Authors: | Ochoa, J.M, Sawaya, M.R, Nguyen, V.N, Duilio, C, Yeates, T.O. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Symmetry breaking and structural polymorphism in a bacterial microcompartment shell protein for choline utilization.
Protein Sci., 29, 2020
|
|
6XPJ
 
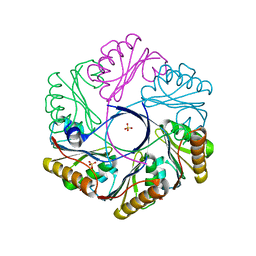 | | CutR flat hexamer, form 2 | | Descriptor: | Ethanolamine utilization protein EutS, SULFATE ION | | Authors: | Ochoa, J.M, Sawaya, M.R, Nguyen, V.N, Duilio, C, Yeates, T.O. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Symmetry breaking and structural polymorphism in a bacterial microcompartment shell protein for choline utilization.
Protein Sci., 29, 2020
|
|
8V9O
 
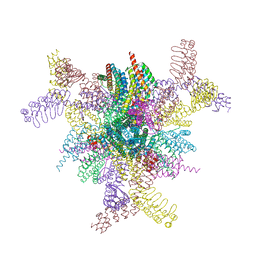 | | Imaging scaffold engineered to bind the therapeutic protein target BARD1 | | Descriptor: | CALCIUM ION, Tetrahedral Nanocage Cage Component Fused to Anti-BARD1 Darpin, Tetrahedral Nanocage Cage, ... | | Authors: | Agdanowski, M.P, Castells-Graells, R, Sawaya, M.R, Yeates, T.O, Arbing, M.A. | | Deposit date: | 2023-12-08 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | X-ray crystal structure of a designed rigidified imaging scaffold in the ligand-free conformation.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
3WIS
 
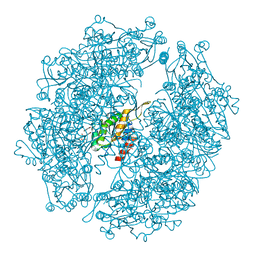 | | Crystal structure of Burkholderia xenovorans DmrB in complex with FMN: A Cubic Protein Cage for Redox Transfer | | Descriptor: | FLAVIN MONONUCLEOTIDE, Putative dihydromethanopterin reductase (AfpA), SULFATE ION | | Authors: | Bobik, T.A, Cascio, D, Jorda, J, McNamara, D.E, Bustos, C, Wang, T.C, Rasche, M.E, Yeates, T.O. | | Deposit date: | 2013-09-25 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure of dihydromethanopterin reductase, a cubic protein cage for redox transfer
J.Biol.Chem., 289, 2014
|
|
5DRK
 
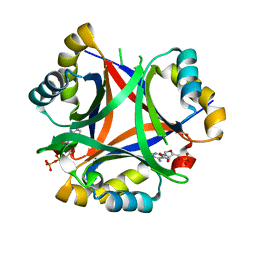 | | 2.3 Angstrom Structure of CPII, a nitrogen regulatory PII-like protein from Thiomonas intermedia K12, bound to ADP, AMP and bicarbonate. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, ... | | Authors: | Wheatley, N.M, Ngo, J, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2015-09-15 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A PII-Like Protein Regulated by Bicarbonate: Structural and Biochemical Studies of the Carboxysome-Associated CPII Protein.
J.Mol.Biol., 428, 2016
|
|
5DS7
 
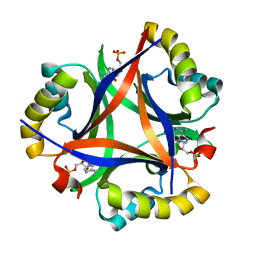 | | 2.0 A Structure of CPII, a nitrogen regulatory PII-like protein from Thiomonas intermedia K12, bound AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Nitrogen regulatory protein P-II | | Authors: | Wheatley, N.M, Ngo, J, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2015-09-17 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A PII-Like Protein Regulated by Bicarbonate: Structural and Biochemical Studies of the Carboxysome-Associated CPII Protein.
J.Mol.Biol., 428, 2016
|
|
1JG1
 
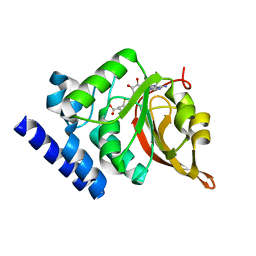 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with S-ADENOSYL-L-HOMOCYSTEINE | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, protein-L-isoaspartate O-methyltransferase | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1JG4
 
 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with S-adenosylmethionine | | Descriptor: | S-ADENOSYLMETHIONINE, protein-L-isoaspartate O-methyltransferase | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1JG3
 
 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with adenosine & VYP(ISP)HA substrate | | Descriptor: | ADENOSINE, CHLORIDE ION, SODIUM ION, ... | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
2B5O
 
 | | ferredoxin-NADP reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, SULFATE ION | | Authors: | Sawaya, M.R, Kerfeld, C.A, Gomez-Lojero, C, Krogmann, D, Bryant, D.A, Yeates, T.O. | | Deposit date: | 2005-09-29 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Crystal Structure of Ferredoxin-NADP reductase from Synechococcus sp. (PCC 7002)
To be Published
|
|
2EWH
 
 | | Carboxysome protein CsoS1A from Halothiobacillus neapolitanus | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Major carboxysome shell protein 1A | | Authors: | Tsai, Y, Sawaya, M.R, Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 2005-11-03 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis of CsoS1A and the Protein Shell of the Halothiobacillus neapolitanus Carboxysome.
Plos Biol., 5, 2007
|
|
1CYI
 
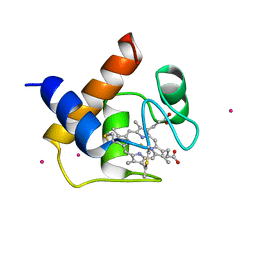 | | CYTOCHROME C6 | | Descriptor: | CADMIUM ION, CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 1995-05-09 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of chloroplast cytochrome c6 at 1.9 A resolution: evidence for functional oligomerization.
J.Mol.Biol., 250, 1995
|
|
1F1C
 
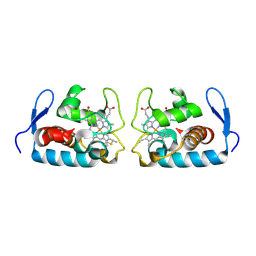 | | CRYSTAL STRUCTURE OF CYTOCHROME C549 | | Descriptor: | CYTOCHROME C549, HEME C | | Authors: | Kerfeld, C.A, Sawaya, M.R, Yeates, T.O, Krogmann, D.W. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
1DOF
 
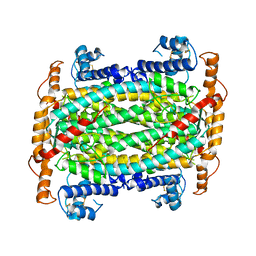 | | THE CRYSTAL STRUCTURE OF ADENYLOSUCCINATE LYASE FROM PYROBACULUM AEROPHILUM: INSIGHTS INTO THERMAL STABILITY AND HUMAN PATHOLOGY | | Descriptor: | ADENYLOSUCCINATE LYASE | | Authors: | Toth, E.A, Yeates, T.O, Goedken, E, Dixon, J.E, Marqusee, S. | | Deposit date: | 1999-12-20 | | Release date: | 2001-01-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of adenylosuccinate lyase from Pyrobaculum aerophilum reveals an intracellular protein with three disulfide bonds.
J.Mol.Biol., 301, 2000
|
|
1KIB
 
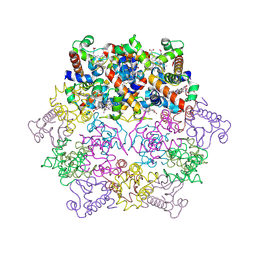 | | cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in the form of an oblate shell | | Descriptor: | HEME C, cytochrome c6 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Krogmann, D, Yeates, T.O. | | Deposit date: | 2001-12-03 | | Release date: | 2002-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in a nearly symmetric shell.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
