1CJ6
 
 | | T11A MUTANT HUMAN LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
1CJ8
 
 | | T40A MUTANT HUMAN LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
1CJ7
 
 | | T11V MUTANT HUMAN LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
1IOF
 
 | | X-RAY CRYSTALLINE STRUCTURES OF PYRROLIDONE CARBOXYL PEPTIDASE FROM A HYPERTHERMOPHILE, PYROCOCCUS FURIOSUS, AND ITS CYS-FREE MUTANT | | Descriptor: | PYRROLIDONE CARBOXYL PEPTIDASE | | Authors: | Tanaka, H, Chinami, M, Ota, M, Tsukihara, T, Yutani, K. | | Deposit date: | 2001-03-09 | | Release date: | 2001-03-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystalline structures of pyrrolidone carboxyl peptidase from a hyperthermophile, Pyrococcus furiosus, and its cys-free mutant.
J.Biochem., 130, 2001
|
|
1CKG
 
 | | T52V MUTANT HUMAN LYSOZYME | | Descriptor: | Lysozyme C | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
2DH6
 
 | | Crystal structure of E. coli Apo-TrpB | | Descriptor: | SULFATE ION, tryptophan synthase beta subunit | | Authors: | Nishio, K, Morimoto, Y, Ogasahara, K, Yasuoka, N, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Large conformational changes in the Escherichia coli tryptophan synthase beta(2) subunit upon pyridoxal 5'-phosphate binding
Febs J., 277, 2010
|
|
2DH5
 
 | | Crystal structure of E. coli Holo-TrpB | | Descriptor: | GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION, ... | | Authors: | Nishio, K, Morimoto, Y, Ogasahara, K, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Large conformational changes in the Escherichia coli tryptophan synthase beta(2) subunit upon pyridoxal 5'-phosphate binding
Febs J., 277, 2010
|
|
2ZFH
 
 | |
2ZQ9
 
 | | Cephalothin acyl-intermediate structure of class a beta-lactamase Toho-1 E166A/R274N/R276N triple mutant | | Descriptor: | 5-METHYLENE-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase Toho-1, CEPHALOTHIN, ... | | Authors: | Nitanai, Y, Shimamura, T, Uchiyama, T, Ishii, Y, Takehira, M, Yutani, K, Matsuzawa, H, Miyano, M. | | Deposit date: | 2008-08-07 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structural Basis of Extend Spectrum Beta-Lactamase in Correlation of Enzymatic Kinetics and Thermal Stability of Acyl-Intermediates
To be Published
|
|
3A1M
 
 | | A fusion protein of a beta helix region of gene product 5 and the foldon region of bacteriophage T4 | | Descriptor: | POTASSIUM ION, chimera of thrombin cleavage site, Tail-associated lysozyme, ... | | Authors: | Yokoi, N, Suzuki, A, Hikage, T, Koshiyama, T, Terauchi, M, Yutani, K, Kanamaru, S, Arisaka, F, Yamane, T, Watanabe, Y, Ueno, T. | | Deposit date: | 2009-04-11 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Construction of Robust Bio-nanotubes using the Controlled Self-Assembly of Component Proteins of Bacteriophage T4
Small, 6, 2010
|
|
2EO8
 
 | |
2E66
 
 | | Crystal Structure Of CutA1 From Pyrococcus Horikoshii OT3, Mutation D60A | | Descriptor: | CHLORIDE ION, Divalent-cation tolerance protein cutA, SODIUM ION | | Authors: | Bagautdinov, B, Sawano, M, Bagautdinova, S, Yutani, K, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-25 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the hyper-thermostability of CutA1
To be Published
|
|
5AVK
 
 | |
5AVJ
 
 | |
1XGO
 
 | |
1XGN
 
 | |
1XGM
 
 | |
1I20
 
 | | MUTANT HUMAN LYSOZYME (A92D) | | Descriptor: | LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
1I1Z
 
 | | MUTANT HUMAN LYSOZYME (Q86D) | | Descriptor: | LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
1I22
 
 | | MUTANT HUMAN LYSOZYME (A83K/Q86D/A92D) | | Descriptor: | CALCIUM ION, LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
3VND
 
 | | Crystal structure of tryptophan synthase alpha-subunit from the psychrophile Shewanella frigidimarina K14-2 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, SULFATE ION, Tryptophan synthase alpha chain | | Authors: | Mitsuya, D, Tanaka, S, Matsumura, H, Takano, K, Urano, N, Ishida, M. | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Strategy for cold adaptation of the tryptophan synthase alpha subunit from the psychrophile Shewanella frigidimarina K14-2: crystal structure and physicochemical properties
J.Biochem., 155, 2014
|
|
3VUQ
 
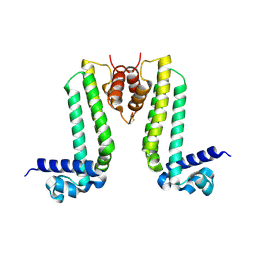 | | Crystal structure of TTHA0167, a transcriptional regulator, TetR/AcrR family from Thermus thermophilus HB8 | | Descriptor: | Transcriptional regulator (TetR/AcrR family) | | Authors: | Agari, Y, Sakamoto, K, Agari, K, Kuramitsu, S, Shinkai, A. | | Deposit date: | 2012-07-04 | | Release date: | 2013-02-27 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and function of a TetR family transcriptional regulator, SbtR, from thermus thermophilus HB8
Proteins, 81, 2013
|
|
1UJP
 
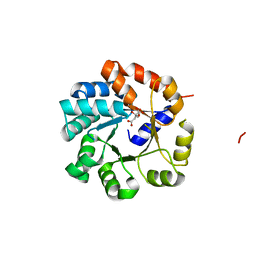 | | Crystal Structure of Tryptophan Synthase A-Subunit From Thermus thermophilus HB8 | | Descriptor: | CITRIC ACID, Tryptophan synthase alpha chain | | Authors: | Asada, Y, Yokoyama, S, Kuramitsu, S, Miyano, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-08 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Stabilization mechanism of the tryptophan synthase alpha-subunit from Thermus thermophilus HB8: X-ray crystallographic analysis and calorimetry.
J.Biochem.(Tokyo), 138, 2005
|
|
1XGS
 
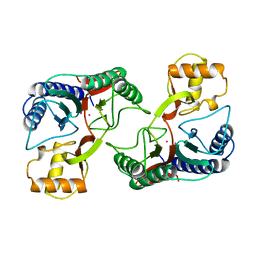 | |
1LZ4
 
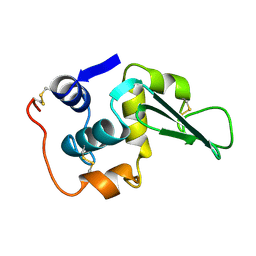 | | ENTHALPIC DESTABILIZATION OF A MUTANT HUMAN LYSOZYME LACKING A DISULFIDE BRIDGE BETWEEN CYSTEINE-77 AND CYSTEINE-95 | | Descriptor: | HUMAN LYSOZYME | | Authors: | Inaka, K, Matsushima, M, Kuroki, R, Taniyama, Y, Kikuchi, M. | | Deposit date: | 1993-02-03 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enthalpic destabilization of a mutant human lysozyme lacking a disulfide bridge between cysteine-77 and cysteine-95.
Biochemistry, 31, 1992
|
|
