1GFC
 
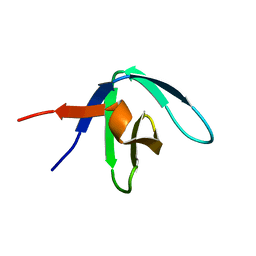 | |
1MKN
 
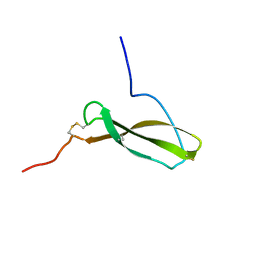 | | N-TERMINAL HALF OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
1WRF
 
 | | Refined solution structure of Der f 2, The Major Mite Allergen from Dermatophagoides farinae | | Descriptor: | Mite group 2 allergen Der f 2 | | Authors: | Ichikawa, S, Takai, T, Inoue, T, Yuuki, T, Okumura, Y, Ogura, K, Inagaki, F, Hatanaka, H. | | Deposit date: | 2004-10-15 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR Study on the Major Mite Allergen Der f 2: Its Refined Tertiary Structure, Epitopes for Monoclonal Antibodies and Characteristics Shared by ML Protein Group Members
J.Biochem.(Tokyo), 137, 2005
|
|
1AHM
 
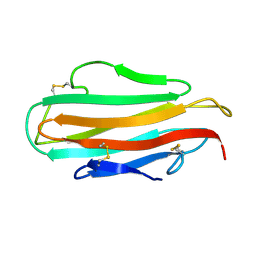 | | DER F 2, THE MAJOR MITE ALLERGEN FROM DERMATOPHAGOIDES FARINAE, NMR, 10 STRUCTURES | | Descriptor: | DER F 2 | | Authors: | Ichikawa, S, Hatanaka, H, Yuuki, T, Iwamoto, N, Ogura, K, Okumura, Y, Inagaki, F. | | Deposit date: | 1997-04-07 | | Release date: | 1998-04-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Der f 2, the major mite allergen for atopic diseases.
J.Biol.Chem., 273, 1998
|
|
1MKC
 
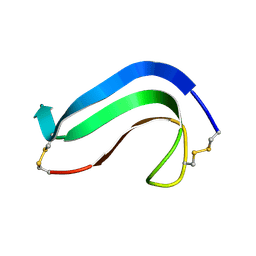 | | C-TERMINAL DOMAIN OF MIDKINE | | Descriptor: | PROTEIN (MIDKINE) | | Authors: | Iwasaki, W, Nagata, K, Hatanaka, H, Ogura, K, Inui, T, Kimura, T, Muramatsu, T, Yoshida, K, Tasumi, M, Inagaki, F. | | Deposit date: | 1999-03-16 | | Release date: | 1999-03-23 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of midkine, a new heparin-binding growth factor.
EMBO J., 16, 1997
|
|
1AHK
 
 | | DER F 2, THE MAJOR MITE ALLERGEN FROM DERMATOPHAGOIDES FARINAE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DER F 2 | | Authors: | Ichikawa, S, Hatanaka, H, Yuuki, T, Iwamoto, N, Ogura, K, Okumura, Y, Inagaki, F. | | Deposit date: | 1997-04-07 | | Release date: | 1998-04-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Der f 2, the major mite allergen for atopic diseases.
J.Biol.Chem., 273, 1998
|
|
2HSP
 
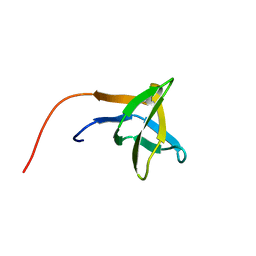 | | SOLUTION STRUCTURE OF THE SH3 DOMAIN OF PHOSPHOLIPASE CGAMMA | | Descriptor: | PHOSPHOLIPASE C-GAMMA (SH3 DOMAIN) | | Authors: | Kohda, D, Hatanaka, H, Odaka, M, Inagaki, F. | | Deposit date: | 1994-06-13 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of phospholipase C-gamma.
Cell(Cambridge,Mass.), 72, 1993
|
|
1IO6
 
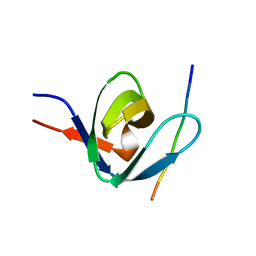 | |
1HSQ
 
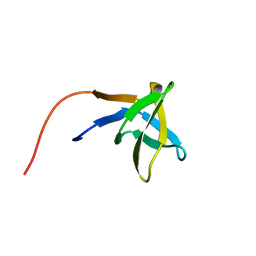 | | SOLUTION STRUCTURE OF THE SH3 DOMAIN OF PHOSPHOLIPASE CGAMMA | | Descriptor: | PHOSPHOLIPASE C-GAMMA (SH3 DOMAIN) | | Authors: | Kohda, D, Hatanaka, H, Odaka, M, Inagaki, F. | | Deposit date: | 1994-06-13 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of phospholipase C-gamma.
Cell(Cambridge,Mass.), 72, 1993
|
|
1D4M
 
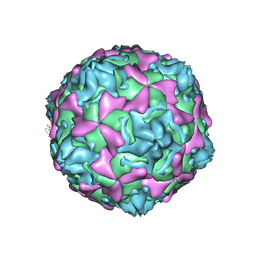 | | THE CRYSTAL STRUCTURE OF COXSACKIEVIRUS A9 TO 2.9 A RESOLUTION | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, MYRISTIC ACID, PROTEIN (COXSACKIEVIRUS A9) | | Authors: | Hendry, E, Hatanaka, H, Fry, E, Smyth, M, Tate, J, Stanway, G, Santti, J, Maaronen, M, Hyypia, T, Stuart, D. | | Deposit date: | 1999-10-04 | | Release date: | 1999-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses.
Structure Fold.Des., 7, 1999
|
|
1IPG
 
 | | SOLUTION STRUCTURE OF THE PB1 DOMAIN OF BEM1P | | Descriptor: | BEM1 PROTEIN | | Authors: | Terasawa, H, Noda, Y, Ito, T, Hatanaka, H, Ichikawa, S, Ogura, K, Sumimoto, H, Inagaki, F. | | Deposit date: | 2001-05-14 | | Release date: | 2001-08-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and ligand recognition of the PB1 domain: a novel protein module binding to the PC motif.
EMBO J., 20, 2001
|
|
1IP9
 
 | | SOLUTION STRUCTURE OF THE PB1 DOMAIN OF BEM1P | | Descriptor: | BEM1 PROTEIN | | Authors: | Terasawa, H, Noda, Y, Ito, T, Hatanaka, H, Ichikawa, S, Ogura, K, Sumimoto, H, Inagaki, F. | | Deposit date: | 2001-04-26 | | Release date: | 2001-08-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and ligand recognition of the PB1 domain: a novel protein module binding to the PC motif.
EMBO J., 20, 2001
|
|
1UHF
 
 | | Solution Structure of the third SH3 domain of human intersectin 2(KIAA1256) | | Descriptor: | INTERSECTIN 2 | | Authors: | Suzuki, S, Hatanaka, H, Koshiba, S, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-03 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the third SH3 domain of human intersectin 2(KIAA1256)
To be Published
|
|
1UHU
 
 | | Solution structure of the retroviral Gag MA-like domain of RIKEN cDNA 3110009E22 | | Descriptor: | product of RIKEN cDNA 3110009E22 | | Authors: | Suzuki, S, Hatanaka, H, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-10 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the retroviral Gag MA-like domain of RIKEN cDNA 3110009E22
To be Published
|
|
1WOT
 
 | | Structure of putative minimal nucleotidyltransferase | | Descriptor: | PUTATIVE MINIMAL NUCLEOTIDYLTRANSFERASE | | Authors: | Suzuki, S, Hatanaka, H, Hondoh, T, Okumura, A, Kuroda, Y, Kuramitsu, S, Shibata, T, Inoue, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-08-25 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of Putative Minimal Nucleotidyltransferase
To be Published
|
|
1J1H
 
 | | Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus | | Descriptor: | Small Protein B | | Authors: | Someya, T, Nameki, N, Hosoi, H, Suzuki, S, Hatanaka, H, Fujii, M, Terada, T, Shirouzu, M, Inoue, Y, Shibata, T, Kuramitsu, S, Yokoyama, S, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-04 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus
FEBS Lett., 535, 2003
|
|
1J03
 
 | | Solution structure of a putative steroid-binding protein from Arabidopsis | | Descriptor: | putative steroid binding protein | | Authors: | Suzuki, S, Hatanaka, H, Kigawa, T, Terada, T, Shirouzu, M, Seki, M, Shinozaki, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-29 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an Arabidopsis homologue of the mammalian membrane-associated progesterone receptor
To be Published
|
|
1HRE
 
 | | SOLUTION STRUCTURE OF THE EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF HEREGULIN-ALPHA, A LIGAND FOR P180ERB4 | | Descriptor: | HEREGULIN ALPHA | | Authors: | Nagata, K, Kohda, D, Hatanaka, H, Ichikawa, S, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-10-15 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epidermal growth factor-like domain of heregulin-alpha, a ligand for p180erbB-4.
EMBO J., 13, 1994
|
|
1K1Z
 
 | | Solution structure of N-terminal SH3 domain mutant(P33G) of murine Vav | | Descriptor: | vav | | Authors: | Ogura, K, Nagata, K, Horiuchi, M, Ebisui, E, Hasuda, T, Yuzawa, S, Nishida, M, Hatanaka, H, Inagaki, F. | | Deposit date: | 2001-09-26 | | Release date: | 2001-10-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of N-terminal SH3 domain of Vav and the recognition site for Grb2 C-terminal SH3 domain
J.BIOMOL.NMR, 22, 2002
|
|
1HRF
 
 | | SOLUTION STRUCTURE OF THE EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF HEREGULIN-ALPHA, A LIGAND FOR P180ERB4 | | Descriptor: | HEREGULIN ALPHA | | Authors: | Nagata, K, Kohda, D, Hatanaka, H, Ichikawa, S, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-10-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epidermal growth factor-like domain of heregulin-alpha, a ligand for p180erbB-4.
EMBO J., 13, 1994
|
|
2CY1
 
 | | Crystal structure of APE1850 | | Descriptor: | NusA protein homolog | | Authors: | Shibata, R, Bessho, Y, Umehara, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization of the archaeal transcription termination factor NusA: a significant decrease in twinning under microgravity conditions
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3A07
 
 | | Crystal Structure of Actinohivin; Potent anti-HIV Protein | | Descriptor: | Actinohivin, SODIUM ION | | Authors: | Tsunoda, M, Suzuki, K, Sagara, T, Takenaka, A. | | Deposit date: | 2009-03-04 | | Release date: | 2009-08-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Mechanism by which the lectin actinohivin blocks HIV infection of target cells
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4XZY
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis | | Descriptor: | GLYCEROL, Peptidase S46 | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2015-02-05 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
4Y01
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis | | Descriptor: | GLYCEROL, Peptidase S46 | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2015-02-05 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
4Y02
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis (Ground) | | Descriptor: | GLYCEROL, POTASSIUM ION, Peptidase S46 | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2015-02-05 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
