1F76
 
 | | ESCHERICHIA COLI DIHYDROOROTATE DEHYDROGENASE | | Descriptor: | Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, FORMIC ACID, ... | | Authors: | Norager, S, Jensen, K.F, Bjornberg, O, Larsen, S. | | Deposit date: | 2000-06-26 | | Release date: | 2002-10-16 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | E. coli Dihydroorotate Dehydrogenase Reveals Structural and Functional Distinction between different classes of
dihydroorotate dehydrogenases
Structure, 10, 2002
|
|
6GKB
 
 | | Iron soak structure of Y40F SynFtn | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Hemmings, A.M, Bradley, J.M. | | Deposit date: | 2018-05-18 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reaction of O2with a diiron protein generates a mixed-valent Fe2+/Fe3+center and peroxide.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6GKA
 
 | | 20 minute Fe2+ soak structure of SynFtn | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin | | Authors: | Hemmings, A.M, Bradley, J.M. | | Deposit date: | 2018-05-18 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Reaction of O2with a diiron protein generates a mixed-valent Fe2+/Fe3+center and peroxide.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8IIO
 
 | | H109Q mutant of uracil DNA glycosylase X | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | Authors: | Aroli, S. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIE
 
 | |
8IIT
 
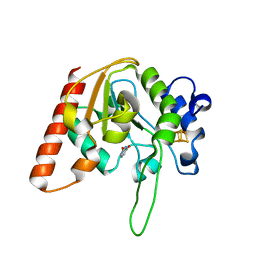 | |
8IIL
 
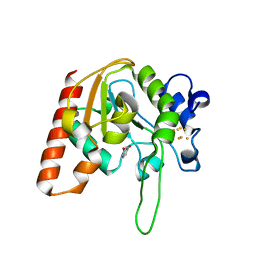 | |
8IIM
 
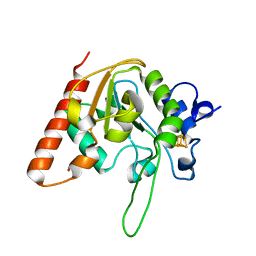 | | H109K mutant of uracil DNA glycosylase X | | Descriptor: | BETA-MERCAPTOETHANOL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | Authors: | Aroli, S. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIH
 
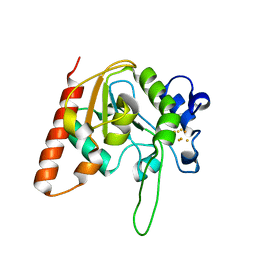 | | H109C mutant of uracil DNA glycosylase X | | Descriptor: | IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | Authors: | Aroli, S. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIQ
 
 | | MsmUdgX H109S/E52N double mutant | | Descriptor: | IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | Authors: | Aroli, S. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIP
 
 | |
8IIS
 
 | | MsmUdgX H109S/R184A double mutant | | Descriptor: | IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | Authors: | Aroli, S. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIJ
 
 | | H109G mutant of uracil DNA glycosylase X | | Descriptor: | BETA-MERCAPTOETHANOL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | Authors: | Aroli, S. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIF
 
 | | H109A mutant of uracil DNA glycosylase X | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | Authors: | Aroli, S. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIN
 
 | |
8IIR
 
 | | MsmUdgX H109S/Q53A double mutant | | Descriptor: | BETA-MERCAPTOETHANOL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | Authors: | Aroli, S. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8III
 
 | |
8IIG
 
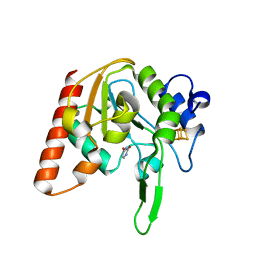 | |
6L1L
 
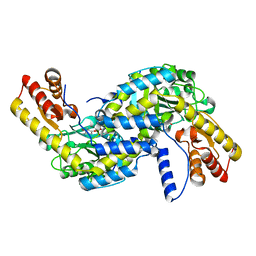 | |
6L1N
 
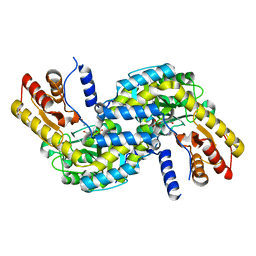 | |
6L1O
 
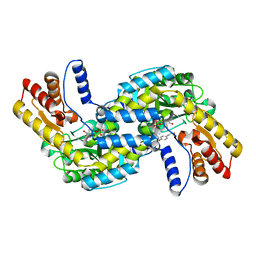 | |
6GKC
 
 | | 2 minute Fe2+ soak structure of SynFtn | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Hemmings, A.M, Bradley, J.M. | | Deposit date: | 2018-05-18 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Reaction of O2with a diiron protein generates a mixed-valent Fe2+/Fe3+center and peroxide.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5EG2
 
 | | SET7/9 N265A in complex with AdoHcy and TAF10 peptide | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, ... | | Authors: | Kroner, G.M, Fick, R.J, Trievel, R.C. | | Deposit date: | 2015-10-26 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Sulfur-Oxygen Chalcogen Bonding Mediates AdoMet Recognition in the Lysine Methyltransferase SET7/9.
Acs Chem.Biol., 11, 2016
|
|
