2OLB
 
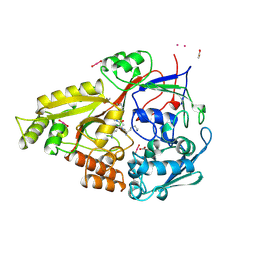 | | OLIGOPEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH TRI-LYSINE | | Descriptor: | ACETATE ION, OLIGO-PEPTIDE BINDING PROTEIN, TRIPEPTIDE LYS-LYS-LYS, ... | | Authors: | Tame, J, Wilkinson, A.J. | | Deposit date: | 1995-09-10 | | Release date: | 1996-01-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of the oligopeptide-binding protein OppA complexed with tripeptide and tetrapeptide ligands.
Structure, 3, 1995
|
|
3DE6
 
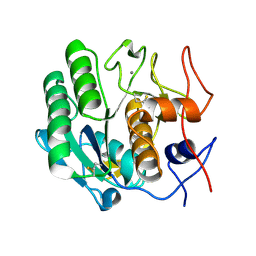 | |
3DE3
 
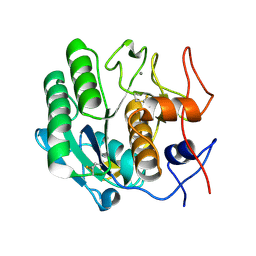 | |
3DDZ
 
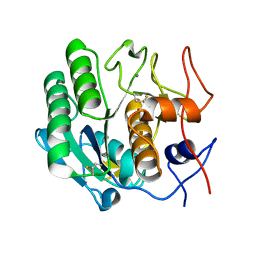 | |
3DE1
 
 | |
3D9Q
 
 | |
3DE0
 
 | |
3DE2
 
 | |
3DE5
 
 | |
3DE7
 
 | |
3DE4
 
 | |
2C6R
 
 | | FE-SOAKED CRYSTAL STRUCTURE OF THE DPS92 FROM DEINOCOCCUS RADIODURANS | | Descriptor: | CHLORIDE ION, DNA-BINDING STRESS RESPONSE PROTEIN, DPS FAMILY, ... | | Authors: | Cuypers, M.G, Romao, C.V, Mitchell, E, Mcsweeney, S. | | Deposit date: | 2005-11-11 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Dps2 from Deinococcus Radiodurans Reveals an Unusual Pore Profile with a Non-Specific Metal Binding Site.
J.Mol.Biol., 371, 2007
|
|
2C2J
 
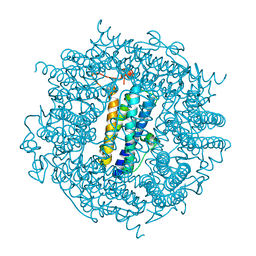 | | Crystal Structure Of The Dps92 From Deinococcus Radiodurans | | Descriptor: | DNA-BINDING STRESS RESPONSE PROTEIN, FE (III) ION, MAGNESIUM ION | | Authors: | Cuypers, M.G, Romao, C.V, Mitchell, E, McSweeney, S. | | Deposit date: | 2005-09-29 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of the Dps2 from Deinococcus Radiodurans Reveals an Unusual Pore Profile with a Non-Specific Metal Binding Site.
J.Mol.Biol., 371, 2007
|
|
3DVS
 
 | |
1REQ
 
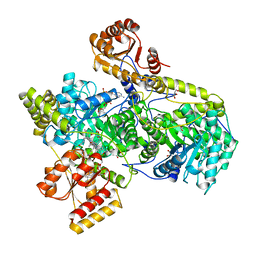 | | METHYLMALONYL-COA MUTASE | | Descriptor: | COBALAMIN, DESULFO-COENZYME A, GLYCEROL, ... | | Authors: | Evans, P.R, Mancia, F. | | Deposit date: | 1996-01-19 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 A resolution.
Structure, 4, 1996
|
|
1AH5
 
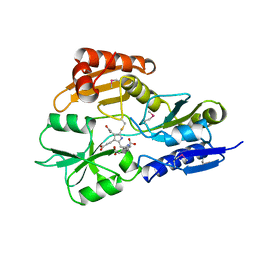 | | REDUCED FORM SELENOMETHIONINE-LABELLED HYDROXYMETHYLBILANE SYNTHASE DETERMINED BY MAD | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, HYDROXYMETHYLBILANE SYNTHASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Harrop, S.J, Cassetta, A. | | Deposit date: | 1997-04-13 | | Release date: | 1997-10-15 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of the structure of seleno-methionine-labelled hydroxymethylbilane synthase in its active form by multi-wavelength anomalous dispersion.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1DJL
 
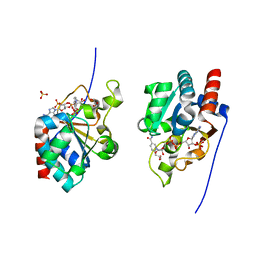 | | THE CRYSTAL STRUCTURE OF HUMAN TRANSHYDROGENASE DOMAIN III WITH BOUND NADP | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | White, S.A, Peak, S.J, Cotton, N.P, Jackson, J.B. | | Deposit date: | 1999-12-03 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The high-resolution structure of the NADP(H)-binding component (dIII) of proton-translocating transhydrogenase from human heart mitochondria.
Structure Fold.Des., 8, 2000
|
|
7M4R
 
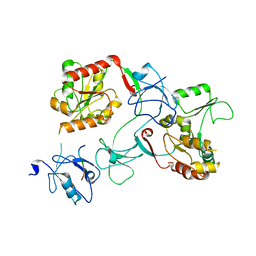 | |
3REQ
 
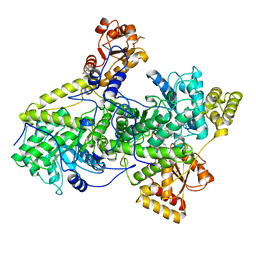 | |
1JEU
 
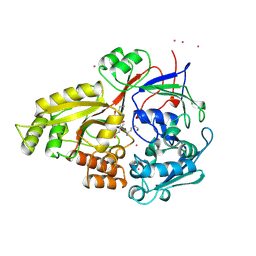 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KEK | | Descriptor: | OLIGO-PEPTIDE BINDING PROTEIN, PEPTIDE LYS GLU LYS, URANYL (VI) ION | | Authors: | Tame, J, Wilkinson, A.J. | | Deposit date: | 1996-07-03 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The role of water in sequence-independent ligand binding by an oligopeptide transporter protein.
Nat.Struct.Biol., 3, 1996
|
|
1JEV
 
 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KWK | | Descriptor: | OLIGO-PEPTIDE BINDING PROTEIN, PEPTIDE LYS TRP LYS, URANYL (VI) ION | | Authors: | Tame, J, Wilkinson, A.J. | | Deposit date: | 1996-07-03 | | Release date: | 1997-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The role of water in sequence-independent ligand binding by an oligopeptide transporter protein.
Nat.Struct.Biol., 3, 1996
|
|
1JET
 
 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KAK | | Descriptor: | OLIGO-PEPTIDE BINDING PROTEIN, PEPTIDE LYS ALA LYS, URANYL (VI) ION | | Authors: | Tame, J, Wilkinson, A.J. | | Deposit date: | 1996-07-03 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The role of water in sequence-independent ligand binding by an oligopeptide transporter protein.
Nat.Struct.Biol., 3, 1996
|
|
1QIF
 
 | |
1QIG
 
 | |
1QIJ
 
 | |
