8W89
 
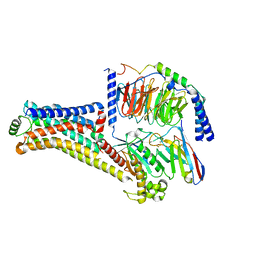 | | Cryo-EM structure of the PEA-bound TAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W87
 
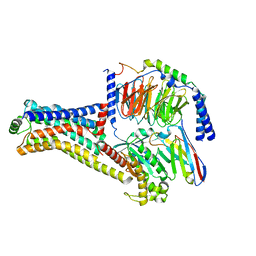 | | Cryo-EM structure of the METH-TAAR1 complex | | Descriptor: | (2S)-N-methyl-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W8A
 
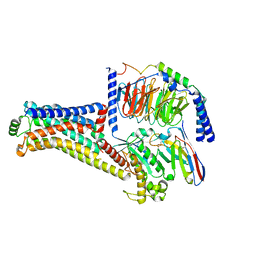 | | Cryo-EM structure of the RO5256390-TAAR1 complex | | Descriptor: | (4S)-4-[(2S)-2-phenylbutyl]-1,3-oxazolidin-2-imine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8WFX
 
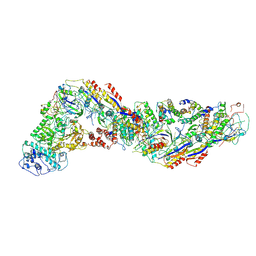 | | Cryo-EM structure of CRISPR-Csm effector complex from Mycobacterium canettii | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | Authors: | Huo, Y, Ma, X, Jiang, T. | | Deposit date: | 2023-09-20 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Type-III-A structure of mycobacteria CRISPR-Csm complexes involving atypical crRNAs.
Int.J.Biol.Macromol., 260, 2024
|
|
8WKT
 
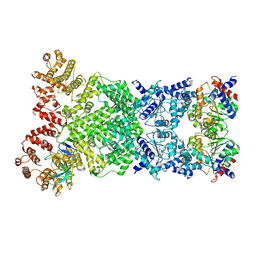 | | Cryo-EM structure of DSR2-DSAD1 complex | | Descriptor: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | Authors: | Gao, A, Huang, J, Zhu, K. | | Deposit date: | 2023-09-28 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Molecular basis of bacterial DSR2 anti-phage defense and viral immune evasion.
Nat Commun, 15, 2024
|
|
8WKX
 
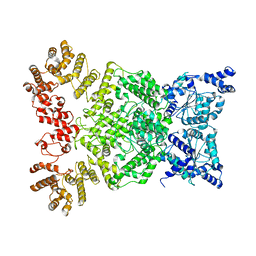 | | Cryo-EM structure of DSR2 | | Descriptor: | SIR2-like domain-containing protein | | Authors: | Gao, A, Huang, J, Zhu, K. | | Deposit date: | 2023-09-28 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Molecular basis of bacterial DSR2 anti-phage defense and viral immune evasion.
Nat Commun, 15, 2024
|
|
8WKS
 
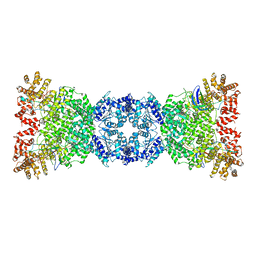 | | Cryo-EM structure of DSR2-TUBE complex | | Descriptor: | SIR2-like domain-containing protein, TUBE | | Authors: | Gao, A, Huang, J, Zhu, K. | | Deposit date: | 2023-09-28 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Molecular basis of bacterial DSR2 anti-phage defense and viral immune evasion.
Nat Commun, 15, 2024
|
|
7VXC
 
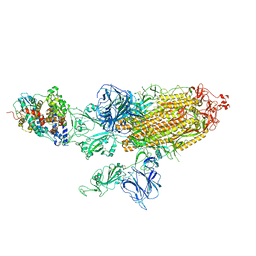 | | SARS-CoV-2 Kappa variant spike protein in C3 state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXI
 
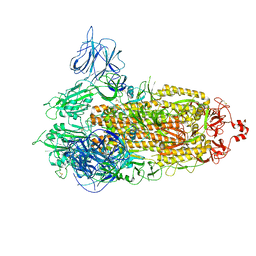 | | SARS-CoV-2 Kappa variant spike protein in transition state | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXK
 
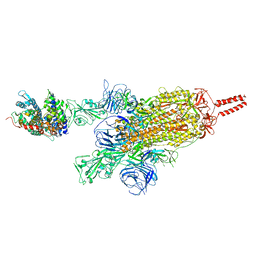 | | SARS-CoV-2 spike protein in complex with ACE2, Beta variant, C2A state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7VXB
 
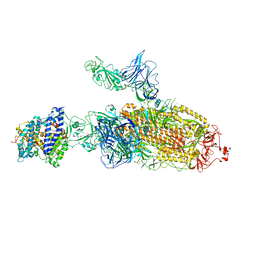 | | SARS-CoV-2 Kappa variant spike protein in C2b state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7W99
 
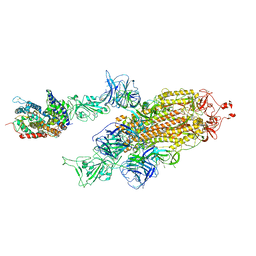 | | SARS-CoV-2 Delta S-ACE2-C2a | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7W94
 
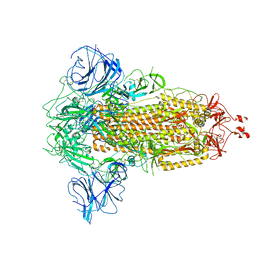 | |
7W9C
 
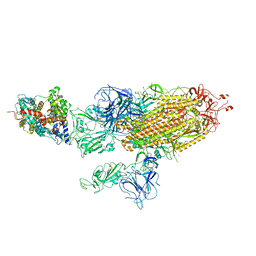 | | SARS-CoV-2 Delta S-ACE2-C3 | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7W98
 
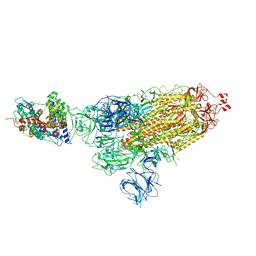 | | SARS-CoV-2 Delta S-ACE2-C1 | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7W9B
 
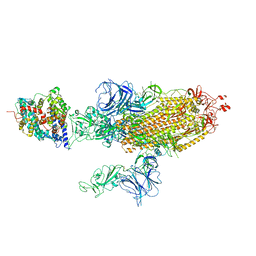 | | SARS-CoV-2 Delta S-ACE2-C2b | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7W92
 
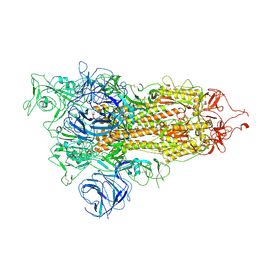 | | Open state of SARS-CoV-2 Delta variant spike protein | | Descriptor: | Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7VXM
 
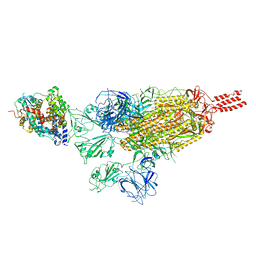 | | SARS-CoV-2 spike protein in complex with ACE2, Beta variant, C3 state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-11-12 | | Release date: | 2022-01-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7W9F
 
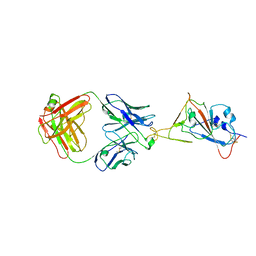 | | SARS-CoV-2 Delta S-RBD-8D3 | | Descriptor: | Spike protein S1, The heavy chain of 8D3, The light chain of 8D3 | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7W9I
 
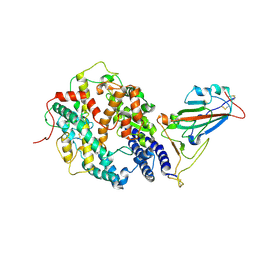 | | SARS-CoV-2 Delta S-RBD-ACE2 | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7W9E
 
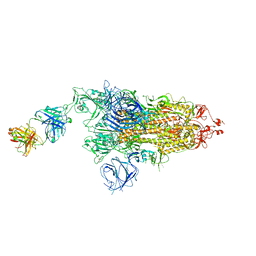 | | SARS-CoV-2 Delta S-8D3 | | Descriptor: | Anti-H5N1 hemagglutinin monoclonal anitbody H5M9 heavy chain, Spike glycoprotein, The light chain of 8D3 fab | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7WD0
 
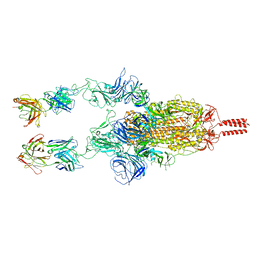 | | SARS-CoV-2 Beta spike in complex with two S5D2 Fabs | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WDF
 
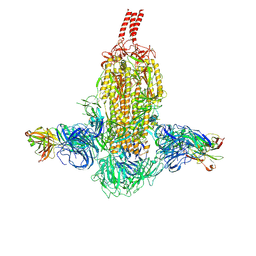 | | SARS-CoV-2 Beta spike in complex with two S3H3 Fabs | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD9
 
 | | SARS-CoV-2 Beta spike in complex with three S3H3 Fabs | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD7
 
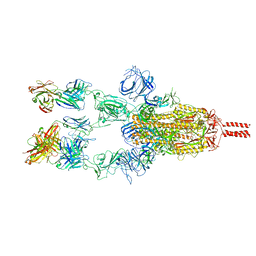 | | SARS-CoV-2 Beta spike in complex with three S5D2 Fabs | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
