7MMB
 
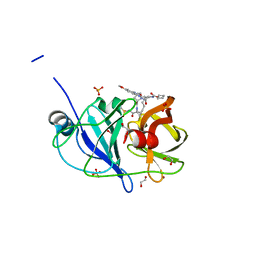 | |
7MMC
 
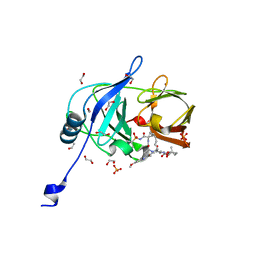 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR01-115 | | Descriptor: | (1-methylcyclopropyl)methyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM4
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR01-115 | | Descriptor: | (1-methylcyclopropyl)methyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MMD
 
 | |
1H2C
 
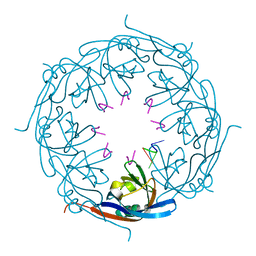 | | Ebola virus matrix protein VP40 N-terminal domain in complex with RNA (High-resolution VP40[55-194] variant). | | Descriptor: | 5'-R(*UP*GP*AP)-3', MATRIX PROTEIN VP40 | | Authors: | Gomis-Ruth, F.X, Dessen, A, Bracher, A, Klenk, H.D, Weissenhorn, W. | | Deposit date: | 2002-08-05 | | Release date: | 2003-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Matrix Protein Vp40 from Ebola Virus Octamerizes Into Pore-Like Structures with Specific RNA Binding Properties
Structure, 11, 2003
|
|
1H2D
 
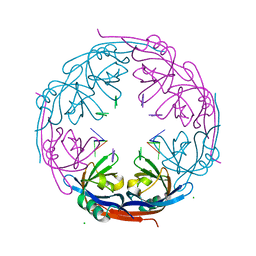 | | Ebola virus matrix protein VP40 N-terminal domain in complex with RNA (Low-resolution VP40[31-212] variant). | | Descriptor: | 5'-R(*UP*GP*AP)-3', CHLORIDE ION, MATRIX PROTEIN VP40 | | Authors: | Gomis-Ruth, F.X, Dessen, A, Bracher, A, Klenk, H.D, Weissenhorn, W. | | Deposit date: | 2002-08-06 | | Release date: | 2003-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Matrix Protein Vp40 from Ebola Virus Octamerizes Into Pore-Like Structures with Specific RNA Binding Properties
Structure, 11, 2003
|
|
4G2M
 
 | |
4G0B
 
 | |
4G22
 
 | |
