2NRT
 
 | |
2NRV
 
 | |
5OKI
 
 | |
5OKC
 
 | |
4ALI
 
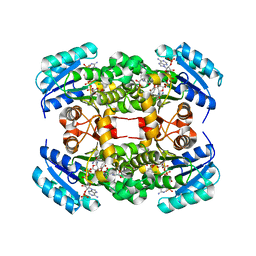 | | Crystal structure of S. aureus FabI in complex with NADP and triclosan (P1) | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | Deposit date: | 2012-03-04 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4ALK
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-ethyl- 2-phenoxyphenol | | Descriptor: | 5-ETHYL-2-PHENOXYPHENOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | Deposit date: | 2012-03-04 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4A15
 
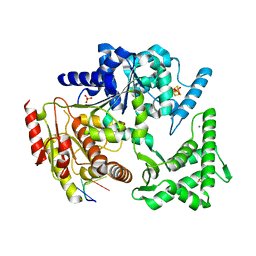 | | Crystal structure of an XPD DNA complex | | Descriptor: | 5'-D(*DTP*AP*CP*GP)-3', ATP-DEPENDENT DNA HELICASE TA0057, CALCIUM ION, ... | | Authors: | Kuper, J, Wolski, S.C, Michels, G, Kisker, C. | | Deposit date: | 2011-09-14 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and Structural Studies of the Nucleotide Excision Repair Helicase Xpd Suggest a Polarity for DNA Translocation.
Embo J., 31, 2011
|
|
4ALN
 
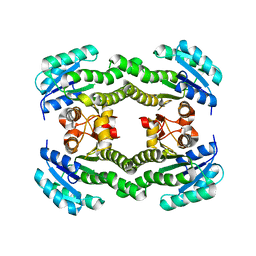 | | Crystal structure of S. aureus FabI (P32) | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH] | | Authors: | Schiebel, J, Kisker, C. | | Deposit date: | 2012-03-04 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4ALJ
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-chloro- 2-phenoxyphenol | | Descriptor: | 5-CHLORO-2-PHENOXYPHENOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | Deposit date: | 2012-03-04 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4ALL
 
 | | Crystal structure of S. aureus FabI in complex with NADP and triclosan (P212121) | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRICLOSAN | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | Deposit date: | 2012-03-04 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4ALM
 
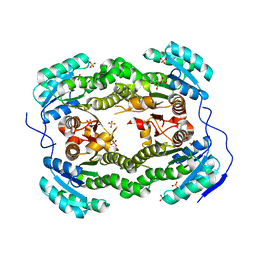 | | Crystal structure of S. aureus FabI (P43212) | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], SULFATE ION | | Authors: | Schiebel, J, Kisker, C. | | Deposit date: | 2012-03-04 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
7AD8
 
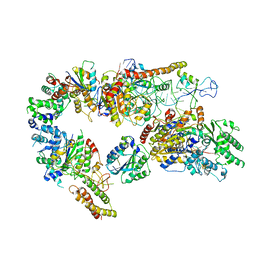 | | Core TFIIH-XPA-DNA complex with modelled p62 subunit | | Descriptor: | DNA (49-MER), DNA repair protein complementing XP-A cells, General transcription and DNA repair factor IIH helicase subunit XPB, ... | | Authors: | Koelmel, W, Kuper, J, Kisker, C. | | Deposit date: | 2020-09-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The TFIIH subunits p44/p62 act as a damage sensor during nucleotide excision repair.
Nucleic Acids Res., 48, 2020
|
|
6EXO
 
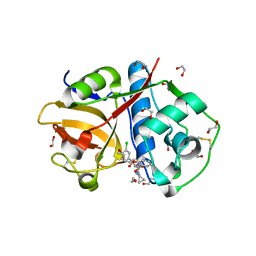 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | Descriptor: | (3~{S},14~{E})-19-chloranyl-~{N}-(1-cyanocyclopropyl)-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(21),6(11),7,9,14,18(22),19-heptaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | Authors: | Dietzel, U, Kisker, C. | | Deposit date: | 2017-11-08 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
6EX8
 
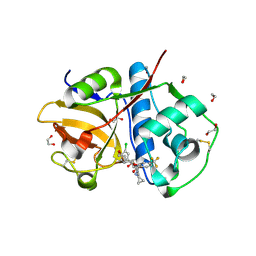 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | Descriptor: | (3~{S})-~{N}-[1-(aminomethyl)cyclopropyl]-19-chloranyl-5-oxidanylidene-9-(trifluoromethyl)-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(20),6(11),7,9,18,21-hexaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | Authors: | Dietzel, U, Kisker, C. | | Deposit date: | 2017-11-07 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
6EXQ
 
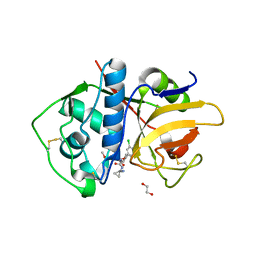 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | Descriptor: | (3~{S})-19-chloranyl-~{N}-(1-cyanocyclopropyl)-8-methoxy-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(20),6(11),7,9,18,21-hexaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | Authors: | Dietzel, U, Kisker, C. | | Deposit date: | 2017-11-08 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
7BMS
 
 | | HEWL in cesium chloride (1.5 M CsCl in crystallization condition and cryo protectant) | | Descriptor: | 1,2-ETHANEDIOL, CESIUM ION, CHLORIDE ION, ... | | Authors: | Koelmel, W, Kuper, J, Kisker, C. | | Deposit date: | 2021-01-20 | | Release date: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cesium based phasing of macromolecules: a general easy to use approach for solving the phase problem.
Sci Rep, 11, 2021
|
|
7BMW
 
 | | p62PH in cesium chloride (0.75 M CsCl in cryo protectant) | | Descriptor: | CESIUM ION, CHLORIDE ION, RNA polymerase II transcription factor B 73 kDa subunit-like protein | | Authors: | Koelmel, W, Kuper, J, Kisker, C. | | Deposit date: | 2021-01-20 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cesium based phasing of macromolecules: a general easy to use approach for solving the phase problem.
Sci Rep, 11, 2021
|
|
7BMO
 
 | | HEWL in cesium chloride (0.25 M CsCl in protein buffer) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Hen egg white lysozyme, ... | | Authors: | Koelmel, W, Kuper, J, Kisker, C. | | Deposit date: | 2021-01-20 | | Release date: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cesium based phasing of macromolecules: a general easy to use approach for solving the phase problem.
Sci Rep, 11, 2021
|
|
7BMP
 
 | | HEWL in cesium chloride (0.25 M CsCl in protein buffer and cryo protectant). | | Descriptor: | 1,2-ETHANEDIOL, CESIUM ION, CHLORIDE ION, ... | | Authors: | Koelmel, W, Kuper, J, Kisker, C. | | Deposit date: | 2021-01-20 | | Release date: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cesium based phasing of macromolecules: a general easy to use approach for solving the phase problem.
Sci Rep, 11, 2021
|
|
7BMU
 
 | | p62PH in cesium chloride (0.25 M CsCl in protein buffer) | | Descriptor: | CESIUM ION, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Koelmel, W, Kuper, J, Kisker, C. | | Deposit date: | 2021-01-20 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cesium based phasing of macromolecules: a general easy to use approach for solving the phase problem.
Sci Rep, 11, 2021
|
|
7BMX
 
 | |
7BMZ
 
 | | p62PH in potassium chloride | | Descriptor: | CHLORIDE ION, POTASSIUM ION, RNA polymerase II transcription factor B 73 kDa subunit-like protein | | Authors: | Koelmel, W, Kuper, J, Kisker, C. | | Deposit date: | 2021-01-20 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cesium based phasing of macromolecules: a general easy to use approach for solving the phase problem.
Sci Rep, 11, 2021
|
|
7BMV
 
 | |
7BMR
 
 | | HEWL in cesium chloride (0.25 M CsCl in protein buffer and 1.71 M CsCl in cryo protectant) | | Descriptor: | 1,2-ETHANEDIOL, CESIUM ION, CHLORIDE ION, ... | | Authors: | Koelmel, W, Kuper, J, Kisker, C. | | Deposit date: | 2021-01-20 | | Release date: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Cesium based phasing of macromolecules: a general easy to use approach for solving the phase problem.
Sci Rep, 11, 2021
|
|
7BMQ
 
 | | HEWL in cesium chloride (1.71 M CsCl in cryo protectant) | | Descriptor: | 1,2-ETHANEDIOL, CESIUM ION, CHLORIDE ION, ... | | Authors: | Koelmel, W, Kuper, J, Kisker, C. | | Deposit date: | 2021-01-20 | | Release date: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Cesium based phasing of macromolecules: a general easy to use approach for solving the phase problem.
Sci Rep, 11, 2021
|
|
