4Z1H
 
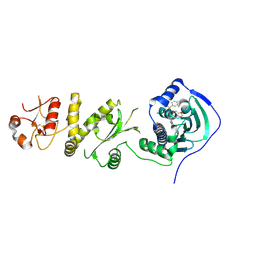 | | Crystal structure of human Trap1 with SMTIN-P01 | | Descriptor: | 8-[(6-iodo-1,3-benzodioxol-5-yl)sulfanyl]-9-[6-(triphenyl-lambda~5~-phosphanyl)hexyl]-9H-purin-6-amine, Heat shock protein 75 kDa, mitochondrial | | Authors: | Lee, C, Park, H.K, Ryu, J.H, Kang, B.H. | | Deposit date: | 2015-03-27 | | Release date: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of a Mitochondria-Targeted Hsp90 Inhibitor Based on the Crystal Structures of Human TRAP1
J.Am.Chem.Soc., 137, 2015
|
|
4Z1I
 
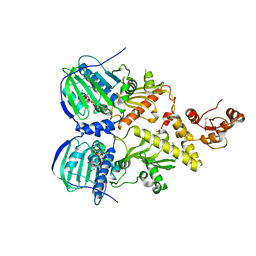 | | Crystal structure of human Trap1 with AMPPNP | | Descriptor: | Heat shock protein 75 kDa, mitochondrial, MAGNESIUM ION, ... | | Authors: | Lee, C, Park, H.K, Ryu, J.H, Kang, B.H. | | Deposit date: | 2015-03-27 | | Release date: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Development of a Mitochondria-Targeted Hsp90 Inhibitor Based on the Crystal Structures of Human TRAP1
J.Am.Chem.Soc., 137, 2015
|
|
7XV2
 
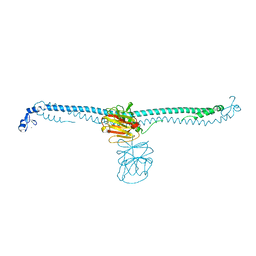 | | TRIM E3 ubiquitin ligase | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2022-05-20 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7XYZ
 
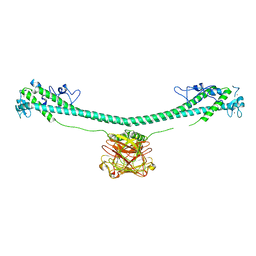 | | TRIM E3 ubiquitin ligase | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2022-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.62 Å) | | Cite: | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7XZ2
 
 | | TRIM E3 ubiquitin ligase | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2022-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7XYY
 
 | | TRIM E3 ubiquitin ligase WT | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2022-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (7.1 Å) | | Cite: | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7XZ1
 
 | | TRIM E3 ubiquitin ligase | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2022-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (5.2 Å) | | Cite: | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6KF3
 
 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
6KBN
 
 | | Crystal structure of Vac8 (del 19-33) bound to Atg13 | | Descriptor: | Autophagy-related protein 13, Vacuolar protein 8 | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-06-25 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Autophagy, 16, 2020
|
|
6KBM
 
 | | Crystal structure of Vac8 bound to Atg13 | | Descriptor: | Autophagy-related protein 13, Vacuolar protein 8 | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-06-25 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Autophagy, 16, 2020
|
|
6KF9
 
 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA (27-MER), DNA (5'-D(P*TP*CP*GP*GP*TP*AP*AP*TP*CP*AP*CP*GP*CP*TP*CP*C)-3'), DNA-directed RNA polymerase subunit, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
6KF4
 
 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
5YI5
 
 | | human ferritin mutant - E-helix deletion | | Descriptor: | Ferritin heavy chain | | Authors: | Lee, S.G, Yoon, H.R, Ahn, B.J, Jeong, H, Hyun, J, Jung, Y, Kim, H. | | Deposit date: | 2017-10-02 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Four-fold Channel-Nicked Human Ferritin Nanocages for Active Drug Loading and pH-Responsive Drug Release
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5VKZ
 
 | | Crystal structure of Mdm12 and combinatorial reconstitution of Mdm12/Mmm1 ERMES complexes for structural studies | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Egea, P.F, AhYoung, A.P, Lu, B, Tan, H.R, Cascio, D. | | Deposit date: | 2017-04-24 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of Mdm12 and combinatorial reconstitution of Mdm12/Mmm1 ERMES complexes for structural studies.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
5YPC
 
 | | p62/SQSTM1 ZZ domain with Phe-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YPB
 
 | | p62/SQSTM1 ZZ domain with His-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YPH
 
 | | p62/SQSTM1 ZZ domain with Ile-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YP8
 
 | | p62/SQSTM1 ZZ domain with Arg-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YPG
 
 | | p62/SQSTM1 ZZ domain with Leu-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YPF
 
 | | p62/SQSTM1 ZZ domain with Trp-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YPE
 
 | | p62/SQSTM1 ZZ domain with Tyr-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YPA
 
 | | p62/SQSTM1 ZZ domain with Lys-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YP7
 
 | | p62/SQSTM1 ZZ domain | | Descriptor: | Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.424 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
6JHK
 
 | |
6K1D
 
 | | Crystal structure of EXD2 exonuclease domain soaked in Mn and GMP | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MANGANESE (II) ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
