4Z2P
 
 | |
4Z28
 
 | | Crystal structure of short hoefavidin biotin complex | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Avidin family, BIOTIN | | Authors: | Livnah, O, Avraham, O. | | Deposit date: | 2015-03-29 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Hoefavidin: A dimeric bacterial avidin with a C-terminal binding tail.
J.Struct.Biol., 191, 2015
|
|
4Z6J
 
 | |
2OZN
 
 | | The Cohesin-Dockerin Complex of NagJ and NagH from Clostridium perfringens | | Descriptor: | CALCIUM ION, CHLORIDE ION, Hyalurononglucosaminidase, ... | | Authors: | Adams, J.J, Boraston, A, Smith, S.P. | | Deposit date: | 2007-02-26 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of Clostridium perfringens toxin complex formation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4Z27
 
 | |
5M0Y
 
 | | Crystal Structure of the CohScaA-XDocCipB type II complex from Clostridium thermocellum at 1.5Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cellulosome anchoring protein cohesin region, ... | | Authors: | Pinheiro, B.A, Bras, J.L, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-10-06 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
3EW2
 
 | | Crystal structure of rhizavidin-biotin complex | | Descriptor: | BIOTIN, rhizavidin | | Authors: | Livnah, O, Meir, A. | | Deposit date: | 2008-10-14 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of rhizavidin: insights into the enigmatic high-affinity interaction of an innate biotin-binding protein dimer.
J.Mol.Biol., 386, 2009
|
|
5M2S
 
 | | R. flavefaciens' third ScaB cohesin in complex with a group 1 dockerin | | Descriptor: | CALCIUM ION, Doc8: Type I dockerin repeat domain from family 9 glycoside hydrolase WP_009982745[Ruminococcus flavefaciens], GLYCEROL, ... | | Authors: | Bule, P, Najmudin, S, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Assembly of Ruminococcus flavefaciens cellulosome revealed by structures of two cohesin-dockerin complexes.
Sci Rep, 7, 2017
|
|
5N5P
 
 | | Crystal structure of Ruminococcus flavefaciens' type III complex containing the fifth cohesin from scaffoldin B and the dockerin from scaffoldin A | | Descriptor: | ACETONITRILE, CALCIUM ION, Putative cellulosomal scaffoldin protein | | Authors: | Bule, P, Carvalho, A.L, Najmudin, S, Fontes, C.M.G.A. | | Deposit date: | 2017-02-14 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Higher order scaffoldin assembly in Ruminococcus flavefaciens cellulosome is coordinated by a discrete cohesin-dockerin interaction.
Sci Rep, 8, 2018
|
|
7P8Z
 
 | | Short wilavidin biotin complex | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, BIOTIN, ... | | Authors: | Avraham, O, Livnah, O. | | Deposit date: | 2021-07-23 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Wilavidin - a novel member of the avidin family that forms unique biotin-binding hexamers.
Febs J., 289, 2022
|
|
7P8Y
 
 | | Short wilavidin apo form | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, PENTAETHYLENE GLYCOL, ... | | Authors: | Avraham, O, Livnah, O. | | Deposit date: | 2021-07-23 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Wilavidin - a novel member of the avidin family that forms unique biotin-binding hexamers.
Febs J., 289, 2022
|
|
7OUQ
 
 | | wilavidin biotin complex | | Descriptor: | BIOTIN, GLYCEROL, wilavidin | | Authors: | Avraham, O, Livnah, O. | | Deposit date: | 2021-06-13 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Wilavidin - a novel member of the avidin family that forms unique biotin-binding hexamers.
Febs J., 289, 2022
|
|
7OUR
 
 | | Wilavidin apo form (P1 form) | | Descriptor: | wilavidin | | Authors: | Avraham, O, Livnah, O. | | Deposit date: | 2021-06-13 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Wilavidin - a novel member of the avidin family that forms unique biotin-binding hexamers.
Febs J., 289, 2022
|
|
5NRK
 
 | | Crystal structure of the sixth cohesin from Acetivibrio cellulolyticus' scaffoldin B in complex with Cel5 dockerin S15I, I16N mutant | | Descriptor: | CALCIUM ION, DocCel5: Type I dockerin repeat domain from A. cellulolyticus family 5 endoglucanase WP_010249057 S15I, I16N mutant, ... | | Authors: | Bule, P, Najmudin, S, Fontes, C.M.G.A, Alves, V.D. | | Deposit date: | 2017-04-24 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-function analyses generate novel specificities to assemble the components of multienzyme bacterial cellulosome complexes.
J. Biol. Chem., 293, 2018
|
|
5NRM
 
 | | Crystal structure of the sixth cohesin from Acetivibrio cellulolyticus' scaffoldin B in complex with Cel5 dockerin S51I, L52N mutant | | Descriptor: | CALCIUM ION, DocCel5: Type I dockerin repeat domain from A. cellulolyticus family 5 endoglucanase WP_010249057 S51I, L52N mutant, ... | | Authors: | Bule, P, Najmudin, S, Fontes, C.M.G.A, Alves, V.D. | | Deposit date: | 2017-04-24 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function analyses generate novel specificities to assemble the components of multienzyme bacterial cellulosome complexes.
J. Biol. Chem., 293, 2018
|
|
5M2O
 
 | | R. flavefaciens' third ScaB cohesin in complex with a group 1 dockerin | | Descriptor: | CALCIUM ION, Group I Dockerin, Putative cellulosomal scaffoldin protein | | Authors: | Bule, P, Najmudin, S, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Assembly of Ruminococcus flavefaciens cellulosome revealed by structures of two cohesin-dockerin complexes.
Sci Rep, 7, 2017
|
|
3T2X
 
 | |
3SZJ
 
 | | Structure of the shwanavidin-biotin complex | | Descriptor: | ACETIC ACID, Avidin/streptavidin, BIOTIN, ... | | Authors: | Livnah, O, Meir, A. | | Deposit date: | 2011-07-19 | | Release date: | 2012-04-11 | | Last modified: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Adaptation of a Thermostable Biotin-binding Protein in a Psychrophilic Environment.
J.Biol.Chem., 287, 2012
|
|
3SZI
 
 | | Structure of apo shwanavidin (P21 form) | | Descriptor: | Avidin/streptavidin, FORMIC ACID | | Authors: | Livnah, O, Meir, A. | | Deposit date: | 2011-07-19 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Adaptation of a Thermostable Biotin-binding Protein in a Psychrophilic Environment.
J.Biol.Chem., 287, 2012
|
|
3T2W
 
 | |
3SZH
 
 | | Crystal structure of apo shwanavidin (P1 form) | | Descriptor: | Avidin/streptavidin | | Authors: | Livnah, O, Meir, A. | | Deposit date: | 2011-07-19 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structural Adaptation of a Thermostable Biotin-binding Protein in a Psychrophilic Environment.
J.Biol.Chem., 287, 2012
|
|
1IJ8
 
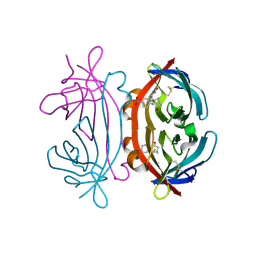 | | CRYSTAL STRUCTURE OF LITE AVIDIN-BNI COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(2-OXO-HEXAHYDRO-THIENO[3,4-D]IMIDAZOL-6-YL)-PENTANOIC ACID (4-NITRO-PHENYL)-AMIDE, AVIDIN | | Authors: | Livnah, O, Huberman, T. | | Deposit date: | 2001-04-25 | | Release date: | 2001-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chicken avidin exhibits pseudo-catalytic properties. Biochemical, structural, and electrostatic consequences.
J.Biol.Chem., 276, 2001
|
|
3ZQW
 
 | | Structure of CBM3b of major scaffoldin subunit ScaA from Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CELLULOSOMAL SCAFFOLDIN, ... | | Authors: | Yaniv, O, Halfon, Y, Lamed, R, Frolow, F. | | Deposit date: | 2011-06-12 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structure of Cbm3B of the Major Scaffoldin Subunit Scaa from Acetivibrio Cellulolyticus
Acta Crystallogr.,Sect.F, 68, 2012
|
|
8AJY
 
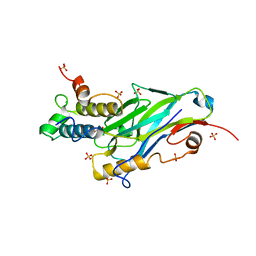 | | Ruminococcus flavefaciens Cohesin-Dockerin structure: dockerin from ScaH adaptor scaffoldin in complex with the cohesin from ScaE anchoring scaffoldin | | Descriptor: | CALCIUM ION, Cell-wall anchoring protein, Dockerin from ScaH, ... | | Authors: | Alves, V.D, Bule, P, Fontes, C.M.G.A, Carvalho, A.L.M, Najmudin, S, Duarte, M. | | Deposit date: | 2022-07-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure-function studies can improve binding affinity of cohesin-dockerin interactions for multi-protein assemblies.
Int.J.Biol.Macromol., 224, 2023
|
|
8ASU
 
 | |
